Abstract
Background
Artemisinin (1) and its derivatives are now being widely used as antimalarial drugs, and they also exhibited good antitumor activities. So there has been much interest in the structural modification of artemisinin and its derivatives because of their effective bioactivities. The microbial transformation is a promising route to obtain artemisinin derivatives. The present study focuses on the microbial transformation of artemisinin by Aspergillus terreus.
Results
During 6 days at 28 °C and 180 rpm, Aspergillus terreus transformed artemisinin to two products. They were identified as 1-deoxyartemisinin (2) and 4α-hydroxy-1-deoxyartemisinin (3) on the basis of their spectroscopic data.
Conclusions
The microbial transformation of artemisinin by Aspergillus terreus was investigated, and two products (1-deoxyartemisinin and 4α-hydroxy-1-deoxyartemisinin) were obtained. This study is the first to report on the microbial transformation of artemisinin by Aspergillus terreus.
Similar content being viewed by others
Background
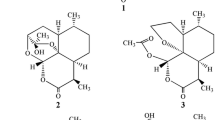
Artemisinin (Fig. 1) 1 (qinghaosu) is a sesquiterpene lactone and its structure was determined by X-ray analysis (Liu et al. 1979). Artemisinin and its derivatives such as dihydroartemisinin, artemether, artesunate, and arteether are now being widely used as antimalaria drugs. In some reports, artemisinin derivatives also exhibited good antitumor activities (Wu et al. 2004; Efferth et al. 2001, 2004; Singh and Lai 2001). There has been much interest in the structural modification of artemisinin and its derivatives because of their effective bioactivities. In this study, we report the microbial transformation of 1 by Aspergillus terreus, and two products were obtained.
Methods
General
1H NMR (nuclear magnetic resonance) and 13C NMR spectra were recorded in CDCl3 (chloroform-d) on a Bruker AV 500 MHz spectrometer. Chemical shifts were reported in ppm (δ), and J values were reported in Hz.
Microorganism
Aspergillus terreus strain ZYL050009 was isolated from soil samples collected from the yew planting base at Guilin, China. The isolate was identified by amplification of the nuclear ribosomal internal transcribed spacer (ITS) region, using the primers ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS 4 (5′-TCC TCC GCT TAT TGA TAT GC-3′) (White et al. 1989). The amplicons were sequenced, and alignments were performed using BLASTN algorithm. Reference sequences with the highest identity were selected and imported into the open source software MEGA 7.0 (Kumar et al. 2016). For the phylogenetic analysis, tree constructions were done with the software MEGA 7.0 using the neighbor-joining method (Hall 2013). Bootstrap analysis was done using 1000-times resampled data.
Medium
All culture and microbial transformation experiments were performed in the following medium: potato infusion is made by boiling 200 g of sliced potatoes in 1 L deionized water for 30 min and then filtering the broth through cheesecloth. Deionized water is added such that the total volume of the suspension is 1 L. 20 g dextrose is then added and the medium is sterilized by autoclaving at 121 °C for 30 min.
Microbial transformation of artemisinin (1) by Aspergillus terreus
Well-developed fungal mycelia were transferred into 250-mL Erlenmeyer flasks containing 60 mL of medium from the surface of agar slants. Cultures were grown for 48 h on a rotary shaker at 28 °C with shaking at 180 rpm, and used to inoculate 51 250-mL shake flasks that contained 60 mL of medium. The cultures were then incubated for 48 h using the same conditions as before. Artemisinin (Mediplantex, Vietnam) was dissolved in acetone (25 mg/mL), filter-sterilized, and 0.4 mL of this solution was added to each flask. A total of 510 mg of artemisinin was transformed. The cultures were incubated for additional 6 days at 28 °C while shaking at 180 rpm. The mycelia were separated by filtration and discarded. The filtrate was extracted three times with an equal volume of ethyl acetate (EtOAc). The extract was evaporated to dryness under vacuum to afford a residue.
Chromatographic conditions
A total of 1.01 g of residue was obtained from the broth. The residue was purified by silica gel column chromatography, using a petroleum ether (60–90 °C)–acetone mobile phase in a gradient mode, eluting with 10–30% acetone.
Results and discussion
We performed DNA sequencing for species identification. We isolated genomic DNA sample from the strain and sequenced the amplified ITS regions. A BLAST search of ITS rDNA sequences available in the GenBank database showed that 583-bp ITS from the strain shared 99% match with A. terreus ATCC 1012 (NR_131276.1), A. terreus strain CCTU1145 (GenBank: KY053120.1), A. terreus isolate NRRL 255 (GenBank: EF669586.1), and A. terreus strain CCTU1009 (GenBank: KY053112.1) (Fig. 2). The isolate was determined as A. terreus.
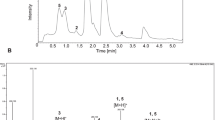
The microbial transformation of artemisinin by A. terreus resulted in 20 mg of 2 (yield 3.9%), 39 mg of 3 (yield 7.6%).
The structures of products were identified on the basis of their spectroscopic data. Data of 1H and 13C NMR spectra of product 2 were in agreement with the reported literatures’ data (Lee et al. 1989; Gaur et al. 2014). So product 2 was identified as 1-deoxyartemisinin (Fig. 1). The comparison of the 1H and 13C NMR data of product 3 with those of 4α-hydroxy-1-deoxyartemisinin (Parshikov et al. 2004; Zhan et al. 2015) was in complete agreement. Therefore, product 3 was confirmed to be 4α-hydroxy-1-deoxyartemisinin (Fig. 1).
1-deoxyartemisinin (2)
Colorless needles (from acetone); 1H-NMR (CDCl3, 500 MHz) δ 0.94 (3H, d, J = 5.6 Hz, Me-14), 0.99 (1H, m, H-8), 1.08 (1H, m, H-7), 1.19 (3H, d, J = 7.2 Hz, Me-15), 1.25 (3H, m, H-5, H-6, H-5a), 1.52 (3H, s, Me-13), 1.63 (1H, m, H-4), 1.78 (2H, m, H-4, H-7), 1.90 (2H, m, H-5, H-8), 2.00 (1H, m, H-8a), 3.18 (1H, m, H-9), 5.69 (1H, s, H-12); 13C NMR (CDCl3, 125 MHz) δ 171.8 (s, C-10), 109.2 (s, C-3), 99.6 (d, C-12), 82.4 (s, C-12a), 44.6 (d, C-5a), 42.4 (d, C-8a), 35.4 (d, C-6), 34.0 (t, C-4), 33.5 (t, C-7), 32.8 (d, C-9), 23.9 (q, C-13), 23.5 (t, C-8), 22.0 (t, C-5), 18.6 (q, C-14), 12.6 (q, C-15).
4α-hydroxy-1-deoxyartemisinin (3)
Colorless needles (from acetone); 1H-NMR (CDCl3, 500 MHz) δ 5.64 (1H, s, H-12), 3.62 (1H, brs, H-4β), 3.20 (1H, m, H-9), 2.06 (1H, m, H-8a), 1.99 (1H, m, H-5α), 1.93 (1H, m, H-8α), 1.82 (1H, m, H-7α), 1.58 (3H, s, Me-13), 1.54 (1H, m, H-5a), 1.50 (1H, m, H-5β), 1.29 (1H, m, H-6), 1.21 (3H, d, J = 7.2 Hz, Me-15), 1.13 (1H, m, H-7β), 1.00 (1H, m, H-8β), 0.93 (3H, d, J = 6.4 Hz, Me-14); 13C NMR (CDCl3, 125 MHz) δ 171.3 (s, C-10), 108.9 (s, C-3), 98.9 (d, C-12), 83.0 (s, C-12a), 69.1 (d, C-4), 42.1 (d, C-8a), 40.6 (d, C-5a), 35.1 (d, C-6), 33.4 (t, C-7), 32.7 (d, C-9), 30.3 (t, C-5), 23.5 (t, C-8), 20.5 (q, C-13), 18.4 (q, C-14), 12.6 (q, C-15).
Although artemisinin is effective against chloroquine-resistant parasites, its toxicities (Kamchonwongpaisan et al. 1997) and low solubility (Lin et al. 1989) in water limit its therapeutical use. The modification of artemisinin has been studied by chemical and biological methods in some reports (Gaur et al. 2014; Parshikov et al. 2004, 2006; Zhan et al. 2002; Goswami et al. 2010; Acton 1999). However, synthesis and semisynthesis of artemisinin derivatives are impossible or impracticable because of the complexity of the artemisinin molecule and the chemical lability of the peroxy ring system. Therefore, microbial transformation is a promising route to obtain artemisinin derivatives. There are many reports on microbial transformation of artemisinin by various microorganisms, such as Aspergillus niger, Rhizopus stolonifer, Cunninghamella elegans, Eurotium amstelodami, Mucor polymorphosporus, Penicillium simplicissimum, Streptomyces griseus (Zhan et al. 2015; Gaur et al. 2014; Goswami et al. 2010; Liu et al. 2006; Parshikov et al. 2004, 2006; Zhan et al. 2002). Here, we first report the microbial transformation of artemisinin by A. terreus.
Conclusions
In this work, we investigated the microbial transformation of artemisinin by A. terreus, and obtained two products, 1-deoxyartemisinin and 4α-hydroxy-1-deoxyartemisinin. This is the first report of microbial transformation of artemisinin by A. terreus.
References
Acton N (1999) Semisynthesis of 3-β-hydroxyartemisinin. J Nat Prod 62:790–793
Efferth T, Dunstan H, Sauerbrey A, Miyachi H, Chitambar CR (2001) The anti-malarial artesunate is also active against cancer. Int J Oncol 18:767–773
Efferth T, Benakis A, Romero MR, Tomicic M, Rauh R, Steinbach D, Häfer R, Stamminger T, Oesch F, Kaina B, Marschall M (2004) Enhancement of cytotoxicity of artemisinins toward cancer cells by ferrous iron. Free Radic Biol Med 37:998–1009
Gaur R, Darokar MP, Ajayakumar PV, Shukla RS, Bhakuni RS (2014) In vitro antimalarial studies of novel artemisinin biotransformed products and its derivatives. Phytochemistry 107:135–140
Goswami A, Saikia PP, Barua NC, Bordoloi M, Yadav A, Bora TC, Gogoi BK, Saxena AK, Suri N, Sharma M (2010) Bio-transformation of artemisinin using soil microbe: direct C-acetoxylation of artemisinin at C-9 by Penicillium simplicissimum. Bioorg Med Chem Lett 20:359–361
Hall BG (2013) Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol 30:1229–1235
Kamchonwongpaisan S, McKeever P, Hossler P, Ziffer H, Meshnick SR (1997) Artemisinin neurotoxicity: neuropathology in rats and mechanistic studies in vitro. Am J Trop Med Hyg 56:7–12
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lee IS, El-Sohly HN, Croon EM, Hufford CD (1989) Microbial metabolism studies of the antimalarial sesquiterpene artemisinin. J Nat Prod 52:337–341
Lin AJ, Lee M, Klayman DL (1989) Antimalarial activity of new water-soluble dihydroartemisinin derivatives. 2. Stereospecificity of the ether side-chain. J Med Chem 32:1249–1252
Liu JM, Ni MY, Fan JF, Tu YY, Wu ZH, Wu YL, Zhou WS (1979) Structure and reaction of arteannuin. Acta Chim Sinica 37:129–143
Liu J-H, Chen Y-G, Yu B-Y, Chen Y-J (2006) A novel ketone derivative of artemisinin biotransformed by Streptomyces griseus ATCC 13273. Bioorg Med Chem Lett 16:1909–1912
Parshikov IA, Muraleedharan KM, Avery MA, Williamson JS (2004) Transformation of artemisinin by Cunninghamella elegans. Appl Microbiol Biotechnol 64:782–786
Parshikov IA, Miriyala B, Muraleedharan KM, Avery MA, Williamson JS (2006) Microbial transformation of artemisinin to 5-hydroxyartemisinin by Eurotium amstelodami and Aspergillus niger. J Ind Microbiol Biotechnol 33:349–352
Singh NP, Lai H (2001) Selective toxicity of dihydroartemisinin and holo-transferrin toward human breast cancer cells. Life Sci 70:49–56
White TJ, Bruns T, Lee SJWT, Taylor JW (1989) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press Inc, San Diego, pp 315–322
Wu GD, Zhou HJ, Wu XH (2004) Apoptosis of human umbilical vein endothelial cells induced by artesunate. Vasc Pharmacol 41:205–212
Zhan J, Zhang Y, Guo H, Han J, Ning L, Guo D (2002) Microbial metabolism of artemisinin by Mucor polymorphosporus and Aspergillus niger. J Nat Prod 65:1693–1695
Zhan Y, Liu H, Wu Y, Wei P, Chen Z, Williamson JS (2015) Biotransformation of artemisinin by Aspergillus niger. Appl Microbiol Biotechnol 99:3443–3446
Authors’ contributions
YZ designed and coordinated the study, performed the data analysis and drafted the manuscript. HY and BZ performed the experiments. All authors read and approved the final manuscript.
Acknowledgments
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets supporting the conclusions of this article are included in the manuscript file.
Ethics approval, consent to participate, and consent for publication
All authors have read and approved to submit it to Bioresources and Bioprocessing.
Funding
This work was supported by Open Funding Project of the State Key Laboratory of Bioreactor Engineering (No. 2015OPEN16), and supported by Guangxi Undergraduate Program for Innovation and Entrepreneurship (No. 201510595170).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Yu, H., Zhu, B. & Zhan, Y. Microbial transformation of artemisinin by Aspergillus terreus . Bioresour. Bioprocess. 4, 33 (2017). https://doi.org/10.1186/s40643-017-0164-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40643-017-0164-6