The importance of GATA factors for development is illustrated by the embryonic lethality of most single GATA knockout mice. Moreover, GATA gene mutations have been described in relation to several human diseases, such as hypoparathyroidism, sensorineural deafness and renal insufficiency (HDR) syndrome, congenital heart diseases (CHDs) and cancer. GATA family members are emerging as potential biomarkers, for instance for the risk prediction of developing acute megalokaryblastic leakemia in Down syndrome and for the detection of colorectal- and breast cancer.
The origin and molecular structure of the GATA family
In vertebrates, six GATA transcription factors have been identified. Based on phylogenetic analysis and tissue expression profiles, the GATA family can be divided into two subfamilies, GATA1/2/3 and GATA4/5/6 (Ref. Reference Simon1). Although in non-vertebrates GATA genes are linked together onto chromosomes, in humans they are segregated onto six distinct chromosomal regions (Table 1), indicating segregation during evolution (Ref. Reference He, Cheng and Zhou2). Most GATA genes encode for several transcripts and protein isoforms. GATA proteins have two zinc finger DNA binding domains, Cys-X2-C-X17-Cys-X2-Cys (ZNI and ZNII), which recognise the sequences (A/T)GATA(A/G) (Fig. 1) (Ref. Reference Lowry and Atchley3). Amongst the six GATA binding proteins, the zinc finger domains are more than 70% conserved, while the sequences of the amino-terminal and carboxyl-terminal domains exhibit lower similarity (Ref. Reference Morrisey4). In non-vertebrates GATA transcription factors have been identified that contain mostly one zinc finger, i.e. in Drosophila melanogaster and Caenorhabditis elegans (Ref. Reference Lowry and Atchley3). The C-terminal zinc finger (ZNII) exists in both vertebrates and non-vertebrates indicating that ZNI was duplicated from ZNII (Ref. Reference He, Cheng and Zhou2).

Figure 1. Overview of GATA1-6 proteins. The GATA proteins are depicted in the upper part of the figure. The GATA proteins are aligned according to the location of the zinc fingers (ZNI and ZNII). The exon boundaries are depicted above the protein structure. For GATA4 the TADI and TADII are shown. In the lower part of the figure the regions around the zinc fingers are enlarged, with the correspondingAA numbers written next to the GATA sequence. Posttranslational modification (post-transciptional modification) sites and disease-associated alterations are marked on top of the corresponding AA. AA, amino acid; TAD, transcriptional activation domains.
Table 1. Molecular features of the human GATA transcription factors

a In the case of multiple transcripts the ensembl transcript ID was chosen, based on the first isoform of the corresponding Uniprot protein sequence.
Tissue-specific roles of GATA factors in development and disease
Haematopoietic system
GATA1/2/3 knockout mice die at the embryonic stage due to haematological abnormalities (Table 2), indicating a pivotal role of these transcription factors in haematopoietic development (Ref. Reference Simon1).
Table 2. Phenotype of GATA knockout mice

Dpc, days post coïtum.
GATA1, the first recognised member of the GATA family, is specifically expressed during haematopoietic development of erythroid, and megakaryocytic cell lineages (Fig. 2) (Ref. Reference Martin11). Loss of GATA1 in mouse embryo-derived stem cells results in a complete lack of primitive erythroid precursor production (Ref. Reference Fujiwara5). Definitive erythroid precursors, on the other hand, are normally produced, but undergo a maturation arrest at the proerythroblast stage followed by apoptosis (Ref. Reference Simon12). Ablation of GATA1 in adult mice also results in a maturation arrest at the same proerythroblast stage (Ref. Reference Yu13). The requirement of the different GATA1 functional domains during primitive and definitive erythropoiesis has been investigated in vivo, showing that both zinc fingers are needed to rescue GATA1 germline mutant mice (Ref. Reference Shimizu14). In haematopoietic stem cells (HSCs), GATA1 gene expression is suppressed, which is indispensable for the maintenance of these stem cells. The mechanism behind this suppression is not fully understood yet. Recently, it was shown that decreased DNA methylation of the GATA1 locus leads to increased GATA2 binding and that increased GATA2 binding results in GATA1 gene transactivation. According to these study results, Takai et al. proposed a mechanism in which GATA1 hypomethylation results in an accessible locus for GATA2 binding which enables transactivation of GATA1 gene expression to initiate erythropoiesis in megakaryo-erythroid progenitors (Ref. Reference Takai15). Loss of GATA1 results in a marked increase of GATA2 expression, indicating not only that GATA2 partially compensates for GATA1 but also that GATA1 suppresses GATA2 transcription during normal erythropoiesis (Ref. Reference Weiss, Keller and Orkin16). This suppression is mediated by the displacement of GATA2 from its upstream enhancer by increasing levels of GATA1 referred to as the ‘GATA switch’ (Ref. Reference Grass17). The combined loss of GATA1 and GATA2 in double-knockout embryos leads to an almost complete absence of primitive erythroid cells, suggesting functional overlap between these transcription factors early in the primitive erythropoiesis (Ref. Reference Fujiwara18).
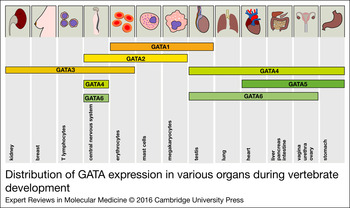
Figure 2. Distribution of GATA expression in various organs during vertebrate development. The expression of all GATA factors is depicted in the corresponding tissues. The distribution of the expression patterns roughly reflects the two GATA subgroups (GATA1/2/3 versus GATA4/5/6).
Requirement of functional GATA1 for haematopoiesis is also observed in several human diseases, such as anaemia, leukaemia and thrombocytopenia (Table 3). Splice site mutations of GATA1 have been found in a family with macrocytic anaemia and in patients with Diamond-Blackfan anaemia (an anaemia characterised by a selective hypoplasia of erythroid cells), resulting in impaired production of the full-length form of the GATA1 protein (Refs Reference Hollanda19, Reference Ludwig20).
Table 3. GATA transcription factors in disease

AMK, acute megakaryoblastic leukaemia; B-ALL, B-cell acute lymphoblastic leukaemia; CHD, congenital heart disease; CML, chronic myeloid leukaemia; DCML, dendritic cell, monocyte, B-lymphocyte and natural killer lymphocyte deficiency; DEL, deletion; DLBCL, diffuse large B-cell lymphoma; DS, Down syndrome; GI, cancer gastrointestinal cance; FS, frameshift; HDR, hypoparathyreoidism, sensorineural deafness and renal disease; INS, insertion; MS, mut missense mutation; MonoMAC, syndrome associated with monocytopenia, B and NK, cell lymphopenia and mycobacterial, fungal and viral infections; NS, mut nonsense mutation; RCC, renal cell carcinoma; SNP, single nucleotide polymorphism; T-ALL, T-cell acute lymphoblastic leukaemia; TMD, transient myeoloproliferative disorder; UCC, urothelial cell carcinoma; XLT, X-linked thrombocytopenia; XLTT, X-linked thrombocytopenia with thalassemia.
Conditional megakaryocytic lineage specific GATA1 knockout mice show excessive marrow megakaryocyte proliferation whereas the platelet numbers are decreased. The maturation of these hyperproliferated megakaryocytes is severely impaired and the produced platelets are structurally and functionally abnormal (Ref. Reference Shivdasani21). Additionally, megakaryocyte-expressed genes with functional GATA1-binding sites (e.g. STAT1) are downregulated in GATA1−/− megakaryocytes (Ref. Reference Huang22). Loss of GATA1 leads to overexpression of GATA2 in megakaryocytes. However GATA1-deficient megakaryocytes still show abnormal megakaryocytic proliferation and differentiation, establishing no functional redundancy of these transcription factors in megakaryopoiesis (Ref. Reference Muntean and Crispino23). In contrast to erythropoiesis, GATA2 remains to be expressed after the GATA switch in late megakaryopoiesis, suggesting a divergent function for both GATA proteins (Ref. Reference Pimkin24).
Children with trisomy 21 are at risk of developing leukaemia, in particular acute megakaryoblastic leukaemia (AMKL). Nearly all Down syndrome patients with AMKL harbour somatic mutations in the GATA1 gene (Table 3) (Ref. Reference Wechsler25), predominantly leading to an N-terminal truncated ‘short’ GATA1 protein (GATA1s) (Ref. Reference Calligaris26). Inadequate GATA1 mediated repression of specific oncogenic factors contributes to megakaryocytic abnormalities (Ref. Reference Li27). Analysis of Down syndrome children with transient myeloproliferative disorder (TMD), which is considered a potential precursor to AMKL, also revealed GATA1 mutations (Ref. Reference Groet28). Noticeable the GATA1 mutation in TMD and subsequent AMKL is identical, suggesting that GATA1 mutations are early events in the development of AMKL in trisomy 21-children (Ref. Reference Hitzler29). Not all TMD Down syndrome neonates with a GATA1 mutation progress to AMKL, indicating the need for more molecular events contributing to the pathogenesis of AMKL. Recently, Yoshida et al. reported newly acquired driver mutations, which lead to the development from TMD to Down syndrome-AMKL (Refs Reference Yoshida30, Reference Nikolaev31).
The mechanism behind the leukaemogenesis remains elusive. Based on mutational spectrum analysis of the GATA1 locus in Down syndrome AMKL, Cabelof et al. hypothesised that increased oxidative stress because of trisomy 21, uracil accumulation and reduced DNA repair together driving leukaemogenesis in Down syndrome (Ref. Reference Cabelof32). Recently it was shown that GATA1 mutations protect megakaryocytes from activated AKT-induced apoptosis (Ref. Reference Stankiewicz and Crispino33). Additionally, trisomy 21 itself increases HSC frequency, clonogenicity and megakaryocyte-erythroid output with associated megakaryocyte-erythroid progenitor expansion (Refs Reference Roy34, Reference Chou35, Reference Maclean36). Another hypothesis is that upregulation of runt-related transcription factor 1 (RUNX1), which physically interacts with GATA1, due to trisomy 21 leads to the induction of GATA1 transcription during embryogenesis, thereby leading to transcription-associated mutagenesis (Ref. Reference Satge37). Recently it is shown that loss of type I interferon (IFN) signalling contributes to GATA1s-induced megakaryocyte hyperproliferation, suggesting AMKL-treatment with IFN-α administration (Ref. Reference Woo38).
GATA1 mutations are also detected in a specific form of X-linked hereditary thrombocytopenia and are described with and without thalassemia (Table 3 and Supplemental Table 1). Hereditary thrombocytopenia without thalassemia has been associated with GATA1 missense mutations that are located in the N-terminal zinc finger region. These mutations lead to loss or inhibition of GATA1 interaction with friend-of-GATA(FOG)1-cofactor (Ref. Reference Nichols39). The degree of disrupted GATA1–FOG1 interaction depends on the mutation, explaining different clinical presentations (Ref. Reference Freson40). The only GATA1 mutation reported in hereditary X-linked thrombocytopenia with thalassemia is the missense mutation R216Q which is located in the DNA binding surface of the GATA1 N-terminal zinc finger and results in reduced DNA binding rather than affecting GATA1–FOG1 interaction (Ref. Reference Yu41).
In vertebrates, GATA2 is expressed in haematopoietic progenitor cells (HPCs), early erythroid cells, mast cells and megakaryocytes, closely resembling the cellular distribution of GATA1 (Fig. 2). A deficit in primitive erythropoiesis is apparent in GATA2−/− mice since the total number of blood cells during embryonic development is markedly reduced, leading to lethality because of severe anaemia (Table 2) (Ref. Reference Tsai6). In GATA2+/− mice haematopoietic defects are seen within HSCs and granulocyte-macrophage progenitor cells. Moreover, the loss of GATA2 in adult mice leads to profound abnormalities in definitive haematopoiesis, also directing to a defect at the level of HSCs (Refs Reference Tsai6, Reference Rodrigues42, Reference Ling43). The function of GATA2 in haematopoietic development has recently been reviewed by Bresnick et al. (Ref. Reference Bresnick44), describing GATA2 as one of the key components establishing the transcriptional program for early haematopoietic development.
Two different GATA2 alterations have been reported in patients with chronic myeloid leukemia (CML) during blast crisis formation (Table 3). In contrast to the in-frame deletion Δ341-346, which leads to decreased transcriptional activation, GATA2 L359V is a gain-of-function mutation and leads to increased DNA binding. Transduction of GATA2 L359V (in vitro and in vivo) resulted in disturbed myelomonocytic differentiation/proliferation, suggesting GATA2 mutations are involved in the acute myeloid transformation of CML (Ref. Reference Zhang45).
GATA2 gene mutations that predisposed to myelodysplastic syndrome (MDS) and acute myeloid leukaemia (AML) were reported (Supplemental Table 1). This occurred either in the absence (non-syndromic) or presence of certain syndromes, including Emberger syndrome and monoMAC syndrome (Ref. Reference Hahn46). Most mutations affect the C-terminal zinc finger or result in N-terminal frameshift mutations (Ref. Reference Hyde and Liu47).
Similar expression patterns of GATA1, GATA2 and GATA3 in human, murine and avian erythroid cells indicate a conserved role for these GATA transcription factors in vertebrate erythropoiesis (Ref. Reference Leonard, Lim and Engel48). Beyond its expression in erythroid lineages, GATA3 is also expressed in T lymphocytes (Ref. Reference Ho49). During haematopoiesis vertebrate GATA3 is expressed in HSCs and in developing T lymphocytes. Murine GATA3−/− embryos are predominantly affected during definitive haematopoiesis in the fetal liver. Although later than GATA2−/− mice, these embryos appear also anaemic and die in utero, probably owing to massive internal bleeding (Table 2) (Ref. Reference Pandolfi7). Frelin et al. demonstrated that GATA3 regulates the self-renewal and differentiation of bone marrow long-term HSCs (Ref. Reference Frelin50). During embryogenesis, GATA3 deficiency leads to a marked reduction in the production of HSCs in the aorta-gonads-mesonephros region. It was shown that GATA3 regulates HSC emergence during embryogenenis via the production of catecholamines linking the haematopoietic system development to the development of the sympathetic nervous system (SNS) (Ref. Reference Fitch51).
In T cell development, GATA3 has a pivotal role from the generation of early T lineage progenitors to CD4+ specification [as reviewed in (Ref. Reference Hosoya, Maillard and Engel52)]. During antigen presentation by specialised antigen-presenting cells, the TCR is stimulated, thereby driving differentiation from peripheral naïve CD4+ T cells towards T helper cell type 1(TH1) or 2 (TH2). GATA3 expression in differentiating TH2 cells is mediated by different pathways as clearly reviewed in Ho et al. (Ref. Reference Ho, Tai and Pai53). GATA3 and STAT6 in TH2 lineage account for lineage specific expression of T cell lincRNAs. At the moment, the function of lincRNAs during T cell development and differentiation is under investigation (Ref. Reference Hu54). An essential function for GATA3 beyond TH2 differentiation is also described demonstrating GATA3 controls proliferation and maintenance of mature T cells (Ref. Reference Wang55).
GATA3 dysregulation is described in leukaemia. Together with T-cell acute lymphocytic leukemia 1 (TAL1) and RUNX1, GATA3 forms an autoregulatory loop that positively regulates the v-myb avian myeloblastosis viral oncogene (MYB) oncogene, which in turn controls the gene expression program in T-cell acute lymphoblastic leukaemia (T-ALL) (Ref. Reference Sanda56). Thereby, whole-genome sequencing of patients with early T-cell precursor ALL, an aggressive subtype of T-ALL, revealed GATA3 inactivating mutations (Supplemental Table 1) (Ref. Reference Zhang57).
In summary, GATA1/2/3 are essential regulators in the development of erythroid and megakaryocytic cell lineages and in the molecular pathogenesis of different haematopoietic diseases.
Cardiovascular system
The mesoderm gives rise to numerous organs, including the heart and genitourinary tract. GATA4/5/6 proteins are expressed in the mesodermal precursors that develop into the heart (Ref. Reference Brewer and Pizzey58).
GATA4 is one of the earliest transcription factors expressed in developing cardiac cells, already detectable in murine precardiac splanchnic mesoderm and associated endoderm (Ref. Reference Kuo8). GATA4−/− mice display severe defects in ventral foregut closure and heart morphogenesis, resulting in embryonic lethality at embryonic day 8 (Table 2). These deformities result from a general loss in ventral folding throughout the embryo and implicate GATA4 requirement for the migration or folding morphogenesis of the precardiogenic splanchic mesodermal cells (Ref. Reference Kuo8). Mice harbouring a knock-in mutation that abrogates the interaction with FOG-cofactors (GATA4Ki/Ki) lack coronary vessels (Ref. Reference Crispino59). In addition, murine GATA4 regulates cardiac angiogenesis by inducing angiogenic factors such as VEGF, facilitating compensation following injury (Ref. Reference Heineke60). Yamak et al. have suggested that GATA4 and Cyclin D2 are part of a forward reinforcing loop in which Cyclin D2 feeds back to enhance cardiogenic activity of GATA4 through direct interaction. GATA4 mutations that abrogate Cyclin D2 interactions are associated with human CHD (Ref. Reference Yamak61).
A variety of GATA4 mutations have been detected in patients with various forms of CHD such as Tetralogy of Fallot, ventricular septal defect and atrial fibrillation as reviewed by McCulley et al and summarised in Table 3 and Supplemental Table 1 (Ref. Reference McCulley and Black62).
Within the developing heart, GATA5 is expressed in the myocardium as well as in the endocardium and derived endocardial cushions in mouse embryos (Ref. Reference Morrisey63). Depending on how GATA5 is inactivated in several mouse models, different cardiac phenotypes are described. Deletion of both GATA5 isoforms leads to hypoplastic hearts and partially penetrant bicuspid aortic valve formation (Ref. Reference Laforest, Andelfinger and Nemer64). When a GATA5 mutant allele was established that lacked the two zinc finger domains, cardiovascular defects were only detectable in a GATA4+/− background (Ref. Reference Singh65). Although little is known about GATA5 in human heart conditions, three heterozygous GATA5 mutations have been associated with familial atrial fibrillation (Ref. Reference Yang66) and four heterozygous GATA5 mutations with CHD (Ref. Reference Jiang67).
GATA6 is abundantly expressed in vascular smooth muscle cells during murine embryonic and postnatal development (Ref. Reference Morrisey68). GATA6 −/− mice die at the embryonic stage due to defects of the extra-embryonic endoderm (Table 2) (Ref. Reference Morrisey10). Tissue-specific deletion of GATA6 in neural crest-derived smooth muscle cells results in an interrupted aortic arch and persistent truncus arteriosus (PTA). These results suggest that GATA6 is required for proper patterning of the aortic arch arteries. This phenotype is associated with severely attenuated expression of semaphorin 3C, a signalling molecule critical for both neuronal and vascular patterning (Ref. Reference Lepore69). Other GATA6 target genes, e.g. Wnt2, in vascular smooth muscle cells and cardiac cells have been identified by microarray analysis after transient GATA6 over-expression. Interestingly, GATA6 is also a target of Wnt2 and together they form a feedforward transcriptional loop to regulate posterior cardiac development (Ref. Reference Tian70).
A number of mutations have been described for GATA6 in the aetiology of CHD (Table 3; Supplemental Table 1). For example, two GATA6 mutations were found in patients with PTA disrupting the transcriptional activity of the GATA6 protein on downstream genes involved in the development of the cardiac outflow tract (Ref. Reference Kodo71).
Thus, the GATA4/5/6 transcription factors have closely related functions during cardiovascular development, and defects lead to CHD and other heart conditions.
Gastrointestinal tract
The endoderm gives rise to the respiratory and gastrointestinal tract as well as the associated organs such as pancreas and liver. Differentiation of embryonic stem cells towards the extra-embryonic endoderm can be induced by forced expression of either GATA4 or GATA6 (Ref. Reference Fujikura72). Targeted mutagenesis of GATA4 in mouse embryonic stem cells results in disturbed differentiation of the visceral endoderm, suggesting that GATA4 has a role in yolk sac formation (Ref. Reference Soudais73).
Murine GATA4 is expressed in the proximal but not in the distal small intestine and has an important role in the maintenance of jejunal-ileal identities (Ref. Reference Boudreau74). Furthermore, GATA4 is essential for jejunal functions such as fat and cholesterol absorption (Ref. Reference Battle75). Beuling et al. found that reduction of GATA4 activity in the intestine induces bile acid absorption in the proximal ileum, which can restore bile acid homeostasis in mice with an ileocaecal resection (Ref. Reference Beuling76).
Whereas GATA4 expression is absent from the distal ileum, GATA6 is expressed throughout the entire small intestine. Conditional deletion of GATA6 in the ileum results in a decrease of crypt cell proliferation and numbers of enteroendocrine and Paneth cells, an increase in numbers of goblet-like cells in crypts and altered expression of genes specific to absorptive enterocytes. GATA4/6 factors are therefore required for proliferation, differentiation and gene expression in the small intestine (Ref. Reference Beuling77).
In humans, GATA4 and GATA5 are expressed in normal gastric and colon mucosa (Refs Reference Akiyama78, Reference Wen79). In gastric and colorectal cancer (CRC) these genes are frequently transcriptionally silenced by methylation (Refs Reference Hellebrekers80, Reference Akiyama78). In addition, we reported that GATA4 and GATA5 exhibit tumour suppressive properties in human CRC cells in vitro (Ref. Reference Hellebrekers80). The potential biomarker capacities of GATA4 are discussed below.
Liver and pancreas
In the mouse, the ventral foregut endoderm differentiates to form the parenchymal components of the liver and ventral pancreas. Although GATA4 has an essential function in embryonic liver development, the protein seems to be dispensable in the adult liver function (Refs Reference Watt81, Reference Zheng82). GATA6−/− murine embryos have defects in endoderm differentiation, and show severely attenuated GATA4 expression levels and complete absence of hepatocyte nuclear factor 4 (HNF4) expression in the visceral endoderm, parietal endoderm and liver bud (Ref. Reference Zhao83). HNF4 is a key regulator for complete differentiation of visceral endoderm, hepatocyte differentiation and the epithelial transformation of the liver (Ref. Reference Parviz84). Tetraploid rescue experiments with GATA6 null mice show that GATA6 is a key regulator for liver bud growth and commitment of the endoderm to a hepatic cell fate (Ref. Reference Zhao83).
Development of the ventral pancreas was, in contrast to the dorsal pancreas, impaired in GATA4−/− murine embryos using tetraploid rescue experiments. GATA6−/− embryos show a similar phenotype, although not as severe as that observed in GATA4−/− embryos (Ref. Reference Watt81). In humans, the role of GATA6 in pancreatic development became apparent in a group of patients with pancreatic agenesis, in which Allen et al. identified 15 de novo heterozygous inactivating mutations in GATA6 (Supplemental Table 1). In addition, these patients suffered from CHD, biliary tract abnormalities, gut developmental disorders, neurocognitive abnormalities and other endocrine abnormalities (Ref. Reference Lango Allen85). In contrast to these results, Martinelli et al. described that GATA6 is dispendable for pancreas development. However, GATA6 is essential for acinar differentiation and maintenance of adult exocrine homeostasis in mice (Ref. Reference Martinelli86). An explanation for this contradiction might be the timepoint of GATA6 inactivation which is earlier in agenesis patients compared with the mouse model used by Martinelli et al. Together these data show the need for further research to unravel the role of GATA6 in pancreatic development.
In pancreatic cancer, GATA6 is often overexpressed, which correlates with GATA6 amplification (Table 3) (Ref. Reference Kwei87). Retained GATA6 expression has been shown in gastric, colorectal, esophageal, ovarian and pulmonary cancer cell lines (Refs Reference Akiyama78, Reference Guo88, Reference Guo89, Reference Caslini90). Additionally, intestinal GATA6 expression is higher in proliferating progenitor cells compared with differentiated cells (Ref. Reference Gao91). In primary gastric cancer, the pro-oncogenic effects of GATA6 are recently confirmed, in vitro and in vivo (Ref. Reference Chia92).
Urogenital tract and kidney
GATA1 is abundantly expressed in the Sertoli cells of the testis during murine prepubertal testis development (Fig. 2). GATA1 expression decreases thereafter and is in the adult mouse testis only found in the Sertoli cells during different stages of the spermatogenesis (Ref. Reference Ito93). Surprisingly, Sertoli-specific GATA1 knockout mice show no alterations in testis development, spermatogenesis, male fertility and expression of putative testis-specific GATA1 target genes (Ref. Reference Lindeboom94). Further research has to clarify whether there is a functional redundancy between GATA factors in the testis.
During urogenital development, GATA4 is expressed in somatic ovarian and testicular cell lineages, and is suggested to have an important regulatory role in gonadal gene expression (Fig. 2) (Ref. Reference Viger95). Mouse embryos conditionally deficient in GATA4 show no formation of the genital ridge, the structure which differentiates into either testis or ovary (Ref. Reference Hu, Okumura and Page96). GATA4ki/ki mice and FOG2−/− mice display defects in the gonadogenesis in both sexes (Ref. Reference Tevosian97). SRY (Y chromosome-linked testis-determining gene), MIS (Mullerian inhibiting substance) and SOX9 expression, which is critical for testis formation, are dependent on GATA4 × FOG2 interaction (Ref. Reference Bouma98). Recently, a signalling cascade was suggested describing transduction of the p38 mitogen-activated protein kinase (MAPK) pathway by MAP3K4 and GADD45G which leads to GATA4 phosphorylation and thereby activation. Phosphorylated GATA4 then binds and activates the SRY promoter (Ref. Reference Gierl99).
The GATA4 gene has also been implicated in a disorder of sex development (DSD). A GATA4 mutation, which abrogates the binding with FOG2, was discovered in a family with both CHD and 46,XY DSD (Table 3) (Ref. Reference Lourenco100). The phenotype closely resembles that of the mouse GATA4ki/ki model (Ref. Reference Tevosian97). The data described above indicate that GATA4, in combination with FOG2, is necessary for proper mammalian sex differentiation.
Murine GATA5 is expressed in the urogenital ridge during foetal development (Ref. Reference Morrisey63). GATA5 −/− female mice exhibit abnormalities of the genitourinary tract including malpositioning of the urogenital sinus, vagina and urethra, whereas males are unaffected (Table 2). These defects suggest that early morphogenic movements in the lower genitourinary tract are disrupted in the absence of GATA5. GATA5 and GATA6 are coexpressed in the developing urogenital ridge but do not seem to have entirely overlapping functions during development of the female genitourinary system (Ref. Reference Molkentin9).
GATA6 is expressed during both testicular and ovarian fetal development (Fig. 2) (Ref. Reference Morrisey63). In the developing gonads, GATA4 and GATA6 have overlapping, but distinct expression patterns, which suggest different roles for these transcription factors. In addition, it is also possible that these factors complement each other's functions because GATA4 and GATA6 are expressed in similar cell types in the testis and ovary (Refs Reference Ketola101, Reference Heikinheimo102).
Loss of GATA6 expression has been found in ovarian cancer and has been associated with hypoacetylation of histones H3 and H4 and loss of H3K4me3 at the promoter region (Ref. Reference Caslini90). Downregulation of GATA6 expression results in nuclear deformation and aneuploidy of ovarian surface epithelial cells (Ref. Reference Capo-chichi103). In contrast to other cancers, these data indicate a tumour suppressor role for GATA6 in ovarian cancer. Tumour suppressing activities are also suggested for GATA4 and GATA5 whereas introduction of these genes into ovarian tumour cell lines greatly inhibits cell growth and survival (Ref. Reference Wakana104).
During pronephros formation human GATA3 expression is already detected in the nephric duct (Fig. 2) (Ref. Reference George105). Subsequently, ureter tips and the collecting duct system of the metanephros are formed, which both show GATA3 expression (Ref. Reference Grote106). Inactivation of the murine GATA3 locus results in a morphologically abnormal nephric duct with an aberrant elongation path, loss of ureteric bud and a severe growth disturbance of de mesonephros due to the disturbance of a regulatory cascade consisting of GATA3 with β-catenin as upstream regulator and Ret as downstream target (Ref. Reference Chia107).
In humans, GATA3 haploinsufficiency leads to the HDR syndrome, a rare and complex disease characterised by the combination of HDR, associated with GATA3 mutations (Table 3, Supplemental Table 1) (Ref. Reference Van Esch108). The majority of these mutations leads to loss of DNA binding caused by a disrupted ZnF2, or altered FOG2 interaction and/or DNA binding affinity by a disrupted ZnF1 (Table 3). Most of the HDR probands without GATA3 mutations do not have renal abnormalities and no GATA3 mutations are found in patients with isolated hypoparathyreoidism (Ref. Reference Ali109). This suggests that GATA3 mutations are highly penetrant and result in the HDR phenotype. In addition, GATA3+/− mice show small size parathyroids resulting in failure to correct hypocalcaemia similar to HDR patients (Ref. Reference Grigorieva110). When GATA3 is specifically deleted in the developing inner ear, defective formation of the cochlear prosensory domain and loss of spiral ganglion neurons is shown (Ref. Reference Luo111). However, the exact mechanisms leading to the HDR phenotype remain to be elucidated.
Respiratory tract
The mammalian lung develops from budding of the foregut endoderm, in which both GATA4 and GATA6 are expressed. In vitro analysis of lung development from GATA4ki/ki mice show abnormal lobar development, revealing GATA4 as a candidate for FOG2-mediated early pulmonary development (Ref. Reference Ackerman112). GATA6-regulated Wnt signalling controls the balance between bronchioalveolar stem cell expansion and epithelial differentiation required for both lung development and regeneration after lung injury (Ref. Reference Zhang113).
However, data about defects in GATA factors in lung diseases are scarce. Recently, GATA2 requirement for oncogenic Kras-driven lung tumorigenenis was reported. Moreover, inhibition of GATA2 regulated pathways in mice with KRAS mutant non-small cell lung cancer results in tumour regression (Ref. Reference Kumar114). Finally, a lung cancer susceptibility locus downstream of GATA3 was identified (Ref. Reference Dong115).
Mammary gland
Using GATA3/LacZ knock-in mice, GATA3 expression is observed at the earliest stages of embryonic mammary development (Fig. 2). During puberty GATA3 is expressed in the terminal-end buds and within the adult mammary gland only in luminal epithelial cells. Targeted GATA3 deletion at different stages of the embryonic mammary development showed loss or absence of mammary primordia and nipples (Ref. Reference Asselin-Labat116). Postnatal GATA3 deletion resulted in loss of mammary gland development, and diminished expression of luminal differentiation markers, which indicates an important role of GATA3 in the luminal epithelium (Refs Reference Asselin-Labat116, Reference Kouros-Mehr117). Loss of the oestrogen receptor α (ERα) expression is observed in both GATA3 knock-out mice and FOG-2 knock-out mice (Ref. Reference Kouros-Mehr117). Involvement of GATA3 and ERα in a positive cross-regulatory loop, which has been shown in breast cancer, may be an explanation for these phenomena (Ref. Reference Eeckhoute118). Collectively, these data show that GATA3 is essential during embryonic development as well as the postnatal occurring morphogenesis (Ref. Reference Asselin-Labat116). Furthermore, GATA3 directs luminal differentiation of progenitor cells and is needed for active maintenance of the differentiated luminal phenotype (Ref. Reference Kouros-Mehr117).
The crucial role of GATA3 in the mammary gland is further demonstrated by the observation of GATA3 mutations in ~10% of human breast cancers. The spectrum of somatic mutations is diverse and cluster predominantly in the vicinity of the highly conserved C-terminal second zinc-finger (Table 3; Supplemental Table 1) (Ref. Reference Koboldt119). Restoration of GATA3 in breast cancer cell lines leads to differentiation, suppressed tumour dissemination (Ref. Reference Kouros-Mehr120), slower growth rates and induction of genes involved in luminal cell differentiation (Ref. Reference Usary121). Thereby, GATA3 expression leads to reduced breast tumour outgrowth and inhibits pulmonary metastasis due to repression of metastasis-associated genes (Ref. Reference Dydensborg122). Recently it was described that GATA3 induces miR-29b expression, which in turn represses metastasis by changing tumour microenvironment (Ref. Reference Chou123). Together these data indicate that GATA3 might function as a tumour suppressor gene. In vitro- and in vivo data support this potential tumour suppressor function because loss of GATA3 leads to tumor progression and tumour dissemination in a murine luminal breast cancer model (Ref. Reference Kouros-Mehr120). Prognostic and predictive features of GATA3 as a biomarker in breast cancer are discussed below in the clinical applications section.
Central Nervous System (CNS)
GATA2 is expressed early during CNS development in murine embryos (Fig. 2) (Ref. Reference Zhou124). Despite early lethality of GATA2 −/− embryos (Table 2), several studies show that GATA2 is required for the development of sympathetic neurons (Ref. Reference Tsarovina125), serotonergic hindbrain neurons (Ref. Reference Craven126), GABAergic midbrain neurons (Ref. Reference Kala127), retinorecipient neurons (Ref. Reference Willett and Greene128) and for the generation and cell fate determination of V2b spinal interneurons (Ref. Reference Zhou, Yamamoto and Engel129). GATA2−/− embryos lack both GATA2 and GATA3 expression in the CNS, which indicates dependence of GATA3 expression on functional GATA2 during early differentiation of the neural tube (Ref. Reference Karunaratne130). The expression pattern of GATA3 during brain development is very similar to GATA2. GATA3−/− murine embryos also die early during embryonic development (Table 2) and have severe abnormalities of the brain and spinal cord (Ref. Reference Pandolfi7). Loss of GATA3 results in reduced Th (tyrosine hydroxylase) and Dbh (dopamine β-hydroxylase) transcripts, which consequently leads to noradrenaline deficiency in the SNS. Administration of catecholamine intermediates to pregnant female GATA3+/− mice rescues GATA3−/− murine embryos, thereby partially unraveling the GATA3 loss-induced lethality (Ref. Reference Lim131). A transcriptional network, which includes GATA3 (Ref. Reference Goridis and Rohrer132), is essential for cell survival and differentiation of sympathetic neurons during embryonic development as well as during adult life (Ref. Reference Tsarovina133).
GATA4 is expressed in the embryonic and adult CNS and acts as a negative regulator of astrocyte proliferation and growth (Fig. 2) (Ref. Reference Agnihotri134). In the adult mouse and human, GATA6 is expressed in neurons, astrocytes, choroids plexus epithelium and endothelial cells (Fig. 2) (Ref. Reference Kamnasaran135).
Loss of expression of GATA4 and GATA6 occurs in glioblastoma multiforme (GBM). Both GATA4/6 gene promoters were found to be methylated and for GATA4 also somatic mutations were found (Refs Reference Agnihotri136, Reference Martinez137). Limited evidence indicates that GATA4 regulates apoptosis-related genes in cultured GBM cell lines (Ref. Reference Agnihotri136). GATA6 was identified in a mouse astrocytoma model as a novel tumour suppressor gene. Knockdown of GATA6 expression in RasV12 or p53−/− astrocytes led to acceleration of tumourigenesis. Mutations of GATA6 occur during malignant progression of murine and human astrocytomas (Ref. Reference Kamnasaran135).
Regulation of GATA genes and proteins in disease
Although mainly GATA gene mutations have been described above, chromosomal alterations as well as regulation of GATA genes and proteins on transcriptional and post-transcriptional levels can also contribute to disease development.
Recently it has been shown that combined tet methylcytosine dioxygenase 2 (TET2) and fms related tyrosine kinase 3 (FLT3) mutations regulate epigenetic silencing of GATA2 by promotor hypermethylation in human AML (Ref. Reference Shih138). In clear cell renal cell carcinomas downregulation of GATA3 expression by promoter hypermethylation results in decreased expression of TbetaRIII, a protein with tumour suppressor features, during disease progression (Ref. Reference Cooper139). Presence of suppressive histone (H3K27) trimethylation of GATA3 together with absence of the GATA3 protein in anaplastic large cell lymphoma implicates epigenetical contribution in the pathogenesis of this disease (Ref. Reference Joosten140). Clues about the transcriptional regulation of the GATA4 and GATA6 genes come from a SUMO-specific protease 2 (SENP2) knockout model. These mice have reduced expression of GATA4 and GATA6 and defects in the embryonic heart. In SENP2 deficient embryos sumoylation of CBX4, accumulates and occupies the promoters of GATA4 and GATA6, thereby leading to transcriptional repression (Ref. Reference Kang141).
GATA4 is located at chromosome 8p, a chromosomal locus frequently deleted in multiple tumour types such as colorectal and oesophageal cancer (Refs Reference Derks142, Reference Lin143). Alternatively GATA4 can be downregulated via epigenetic silencing, such as hypoacetylation of histones H3 and H4 (Ref. Reference Caslini90) and promoter CpG island hypermethylation, which has been observed in colorectal, gastric, oesophageal, lung, ovarian and HPV-driven oropharyngeal cancer, in GBM and in diffuse large B-cell lymphoma (Refs Reference Hellebrekers80, Reference Akiyama78, Reference Guo88, Reference Guo89, Reference Wakana104, Reference Agnihotri136, Reference Kostareli144, Reference Pike145). In contrast, GATA4 amplification is recently described in certain gastric cancer which indicates a more oncogenic function (Ref. Reference Chia92). Further studies are needed to unravel the molecular mechanisms of GATA4 amplified in comparison with GATA4 methylated gastric cancers.
GATA5 is located at chromosome 20q13, a locus which is often amplified and methylated in multiple cancer types. No coding sequence mutations in GATA4 and GATA5 have been described so far in colorectal- and breast cancer (Refs Reference Sjoblom146, Reference Wood147). However, promoter methylation of GATA5 might be established in order to downregulate increased gene expression imposed by amplification. Identified post-transcriptional modifications on GATA proteins include acetylation, phosphorylation and methylation (Fig. 1). Protein stability of GATA2 and GATA3 is regulated by phosphorylation and ubiquitilation. Phosphorylation of GATA3 by respectively Cyclin-dependent kinase 1 (CDK1) and CDK2 was required for F-box/WD repeat-containing protein 7 (Fbw)-7 mediated ubiquitilation and degradation and contributed to precise differentiation of HSCs and T-cell lineages (Refs Reference Kitagawa148, Reference Nakajima149). How GATA acetylation influences transcriptional processes has been investigated for GATA1. It turns out that bromodomain protein Brd3 binds to acetylated GATA1 to regulate the chromatin occupancy at erythroid target genes (Ref. Reference Lamonica150). For GATA4 post-transciptional modifications have mainly been studied in the context of hypertrophy of the heart. Activation of GATA4 occurs in part through acetylation by the transcriptional coactivator p300. Takaya et al. identified 4 GATA4 lysine residues that, when mutated, lacked p300-induced acetylation, DNA binding and transcriptional activities (Fig. 1) (Ref. Reference Takaya151). Phosphorylation of p300 by Cdk9 increases the ability of p300 to induce acetylation and DNA binding of GATA4 (Ref. Reference Sunagawa152). Alternatively, phosphorylation of GATA4 on serine 105 is critical for a productive cardiac hypertrophic response to stress stimulation in adult mice (Ref. Reference van Berlo153). Deacetylation of GATA4, and subsequent suppression of transcriptional activation, is mediated by histone deacetylase 2 (HDAC2) and the small homeodomain factor Hopx (Ref. Reference Trivedi154). Recently it was reported that the GATA4 protein is methylated by Polycomb-repressive complex 2 member Ezh2. This reduced the interaction with and acetylation by p300, thereby reducing GATA4's transcriptional activity (Ref Reference He155). Together, this emphasises how important post-transciptional modifications are for the regulation of GATA activity.
Clinical applications of GATA transcription factor alterations
The above mentioned alterations in GATA factors might be applicable as biomarkers for early detection, diagnosis and prediction of prognosis and response to therapy.
Early detection markers
Non-invasive early diagnosis of CRC reduces mortality of this disease (Ref. Reference Hewitson156). We have shown that GATA4 promoter methylation is highly prevalent in CRC, suggesting that methylation is an early event in colorectal carcinogenesis. GATA4 methylation, detected in faecal DNA has potential to be used as a biomarker for improving pre-selection tests for colonoscopy (Ref. Reference Hellebrekers80), especially if the clinical and analytical sensitivity and specificity can be improved by adding additional biomarkers and by introducing sensitive analysis techniques such as for example methylation on beads technology (Ref. Reference Guzzetta157).
Diagnostic markers
The expression of several GATA factors can be helpful in establishing a correct diagnosis. In ovarian cancer loss of GATA4 precedes loss of GATA6 expression and can differentiate between histological subtypes. Loss of both GATA4 and GATA6 expression is found in serous, clear cell and endometrioid ovarian cancer, but their expression can be detected in mucinous carcinomas (Ref. Reference Cai158).
Prognostic markers
As already described above, GATA1 mutations are found in nearly all AMKL patients with Down syndrome and are already detectable in the precursor lesion TMD. In addition, Down syndrome-neonates without GATA1 mutations do not develop AMKL (Refs Reference Pine159, Reference Roberts160). Together, the presence of GATA1 mutations in Down syndrome-children might be a potential prognostic marker for identifying infants at higher risk of developing AMKL (Ref. Reference Roy161). Besides having a clinical value in AMKL, prognostic properties of GATA transcription factors are also described in T-ALL. Inherited genetic GATA3 variants are identified in Philadelphia-like ALL (an ALL subtype with a poor prognosis) and are associated with early treatment response and a higher risk of relapse (Ref. Reference Perez-Andreu162).
GATA3 downregulation has been observed in ER-negative breast cancers and has been described as a strong prognostic indicator of breast cancer. Low GATA3 expression was strongly associated with aggressive disease and poor survival (Ref. Reference Kouros-Mehr117). Vice versa, breast cancers expressing GATA3- and estrogen regulated genes exhibit a good prognosis and have better relapse-free and overall survival (Ref. Reference Oh163). GATA3 has been considered to be a better prognostic marker for disease-free survival than commonly used variables such as ER status (Ref. Reference Mehra164) although conflicting data have been published. However, GATA3 expression is highly correlated with the luminal A subtype which has a relatively favourable outcome compared with luminal B and basal-like subtypes (Ref. Reference Albergaria165). An explanation could be the downregulation of p18INK4C transcription by GATA3 resulting in expansion of luminal progenitor cells thereby favouring the development of luminal type breast cancer (Ref. Reference Pei166).
Recent studies indicate that GATA2 may be a useful biomarker for predicting prognosis in AML. GATA2 mutations are frequent in patients with a biallelic CEBPA mutation and are associated with a better survival (Ref. Reference Fasan167).
In oropharyngeal carcinomas, a methylation signature of 5 gene promoters, including GATA4, correlates with improved survival (Ref. Reference Kostareli144). Eventually, loss of expression of GATA4 in GBM is associated with unfavourable patient survival (Ref. Reference Agnihotri136).
Recently it has been described that low GATA6 expression in lung adenocarcinomas is linked to increased incidence of metastasis and poor outcome (Ref. Reference Cheung168).
Predictive markers
Whole genome sequencing of samples from patients with ER-positive breast cancer, participating in aromatase inhibitor clinical trials identified 18 significantly mutated genes, including GATA3. Mutant GATA3 correlated with suppression of proliferation upon aromatase inhibitor treatment and might therefore be a positive predictive marker for aromatase inhibitor response (Ref. Reference Ellis169).
Re-expression of GATA4 in GBM cells conferred sensitivity to temozolomide, a DNA alkylating agent used in GBM therapy (Ref. Reference Agnihotri136).
Recently, GATA5 methylation was described as a potential predictive marker for patients with high-risk non-muscle-invasive bladder tumours. These patients had a better survival after treatment with Bacillus Calmette-Guérin (BCG) when GATA5 was methylated (Ref. Reference Agundez170).
Therapeutic interventions
For regenerative medicine the generation of functional differentiated cell types is of great therapeutic interest. Since heart disease occurs frequently and the heart has little regenerative capacity after damage, procedures are sought that can transdifferentiate fibroblast into cardiac myocytes. A cocktail of transcription factors, including GATA4 converts cardiac non-myocytes into cardiomyocyte-like cells in vivo, and alleviates cardiac injury (Refs Reference Song171, Reference Qian172). Also in mouse liver engineering experiments GATA4 was one of the essential factors that contributed to the conversion of fibroblasts into functional hepatocyte-like cells (Ref. Reference Huang173). These induced cells were able to restore liver function in half of fumarylacetoacetate-hydrolase-deficient mice. GATA4 is thus one of the pivotal genes that in combination with other transcription factors can be utilised to improve heart and liver function after damage. These promising results are the first steps for bringing regenerative medicine to the clinic. More knowledge of the different GATA protein functions and their downstream target genes is necessary before therapeutic strategies can be developed.
Conclusions and future perspectives
An increasing number of studies are being published, describing expression and function of GATA genes during development in different species.
Causal relationships between aberrations in GATA genes and several human diseases have become apparent. Numerous mutations in the GATA genes have been described above. Many disease-associated mutations are located in and around the Zinc finger regions. As those mutations are not specifically limited to the two Zinc fingers themselves, it is clear that the whole region is important for the proteins to be fully operational. Most likely mutations hinder the correct folding of the proteins and thereby obstruct GATA proteins from binding to their relevant binding partners. The application of next-generation sequencing technologies through whole-genome, whole-exome and whole-transcriptome approaches allows for substantial advances, which is expected to reveal more disease-associated alterations whithin GATA genes.
A better understanding of the regulation of GATA factors on transcriptional, translational and post-translational levels will give more leads to how GATAs can be used as biomarkers. Prospective clinical trials, based on these data, are necessary to determine the translational value of GATA genes as biomarkers.
Supplementary material
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/erm.2016.2









