当前位置:
X-MOL 学术
›
Genes. Immun.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Prioritizing Crohn's disease genes by integrating association signals with gene expression implicates monocyte subsets.
Genes and Immunity ( IF 5 ) Pub Date : 2019-01-29 , DOI: 10.1038/s41435-019-0059-y Kyle Gettler 1 , Mamta Giri 2 , Ephraim Kenigsberg 2 , Jerome Martin 2 , Ling-Shiang Chuang 2 , Nai-Yun Hsu 2 , Lee A Denson 3 , Jeffrey S Hyams 4 , Anne Griffiths 5 , Joshua D Noe 6 , Wallace V Crandall 7 , David R Mack 8 , Richard Kellermayer 9 , Clara Abraham 10 , Gabriel Hoffman 2 , Subra Kugathasan 11, 12 , Judy H Cho 2, 13
Genes and Immunity ( IF 5 ) Pub Date : 2019-01-29 , DOI: 10.1038/s41435-019-0059-y Kyle Gettler 1 , Mamta Giri 2 , Ephraim Kenigsberg 2 , Jerome Martin 2 , Ling-Shiang Chuang 2 , Nai-Yun Hsu 2 , Lee A Denson 3 , Jeffrey S Hyams 4 , Anne Griffiths 5 , Joshua D Noe 6 , Wallace V Crandall 7 , David R Mack 8 , Richard Kellermayer 9 , Clara Abraham 10 , Gabriel Hoffman 2 , Subra Kugathasan 11, 12 , Judy H Cho 2, 13
Affiliation
|
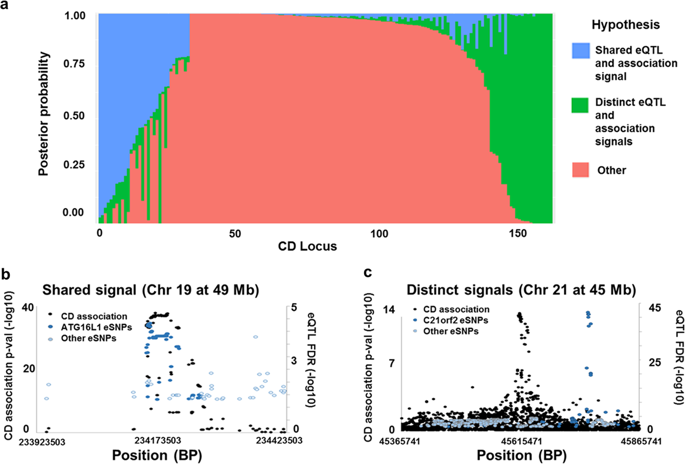
Genome-wide association studies have identified ~170 loci associated with Crohn's disease (CD) and defining which genes drive these association signals is a major challenge. The primary aim of this study was to define which CD locus genes are most likely to be disease related. We developed a gene prioritization regression model (GPRM) by integrating complementary mRNA expression datasets, including bulk RNA-Seq from the terminal ileum of 302 newly diagnosed, untreated CD patients and controls, and in stimulated monocytes. Transcriptome-wide association and co-expression network analyses were performed on the ileal RNA-Seq datasets, identifying 40 genome-wide significant genes. Co-expression network analysis identified a single gene module, which was substantially enriched for CD locus genes and most highly expressed in monocytes. By including expression-based and epigenetic information, we refined likely CD genes to 2.5 prioritized genes per locus from an average of 7.8 total genes. We validated our model structure using cross-validation and our prioritization results by protein-association network analyses, which demonstrated significantly higher CD gene interactions for prioritized compared with non-prioritized genes. Although individual datasets cannot convey all of the information relevant to a disease, combining data from multiple relevant expression-based datasets improves prediction of disease genes and helps to further understanding of disease pathogenesis.
中文翻译:

通过将关联信号与基因表达整合在一起,对克罗恩病基因进行优先排序会涉及单核细胞亚群。
全基因组关联研究已经确定了约170个与克罗恩病(CD)相关的基因座,确定哪些基因驱动这些关联信号是一个重大挑战。这项研究的主要目的是确定哪些CD基因座基因最可能与疾病相关。通过整合互补的mRNA表达数据集,包括来自302名新诊断,未治疗的CD患者和对照以及受刺激的单核细胞末端回肠的大量RNA-Seq,我们开发了基因优先排序回归模型(GPRM)。在回肠RNA-Seq数据集上进行了转录组范围内的关联和共表达网络分析,确定了40个基因组范围内的重要基因。共表达网络分析鉴定了单个基因模块,该模块显着富集了CD基因座基因,并且在单核细胞中表达最高。通过包括基于表达的信息和表观遗传信息,我们将可能的CD基因从每个基因座平均7.8个平均基因中细化为2.5个优先基因。我们使用交叉验证验证了我们的模型结构,并通过蛋白质关联网络分析验证了我们的优先排序结果,与非优先排序的基因相比,这证明了优先排序的CD基因相互作用更高。尽管单个数据集无法传达与疾病相关的所有信息,但将多个基于表达的相关数据集相结合的数据可以改善疾病基因的预测,并有助于进一步了解疾病的发病机理。我们使用交叉验证验证了我们的模型结构,并通过蛋白质关联网络分析验证了我们的优先排序结果,结果表明,与非优先排序的基因相比,优先排序的CD基因相互作用更高。尽管单个数据集无法传达与疾病相关的所有信息,但将多个基于表达的相关数据集相结合的数据可以改善疾病基因的预测,并有助于进一步了解疾病的发病机理。我们使用交叉验证验证了我们的模型结构,并通过蛋白质关联网络分析验证了我们的优先排序结果,结果表明,与非优先排序的基因相比,优先排序的CD基因相互作用更高。尽管单个数据集无法传达与疾病相关的所有信息,但将多个基于表达的相关数据集相结合的数据可以改善疾病基因的预测,并有助于进一步了解疾病的发病机理。
更新日期:2019-01-29
中文翻译:

通过将关联信号与基因表达整合在一起,对克罗恩病基因进行优先排序会涉及单核细胞亚群。
全基因组关联研究已经确定了约170个与克罗恩病(CD)相关的基因座,确定哪些基因驱动这些关联信号是一个重大挑战。这项研究的主要目的是确定哪些CD基因座基因最可能与疾病相关。通过整合互补的mRNA表达数据集,包括来自302名新诊断,未治疗的CD患者和对照以及受刺激的单核细胞末端回肠的大量RNA-Seq,我们开发了基因优先排序回归模型(GPRM)。在回肠RNA-Seq数据集上进行了转录组范围内的关联和共表达网络分析,确定了40个基因组范围内的重要基因。共表达网络分析鉴定了单个基因模块,该模块显着富集了CD基因座基因,并且在单核细胞中表达最高。通过包括基于表达的信息和表观遗传信息,我们将可能的CD基因从每个基因座平均7.8个平均基因中细化为2.5个优先基因。我们使用交叉验证验证了我们的模型结构,并通过蛋白质关联网络分析验证了我们的优先排序结果,与非优先排序的基因相比,这证明了优先排序的CD基因相互作用更高。尽管单个数据集无法传达与疾病相关的所有信息,但将多个基于表达的相关数据集相结合的数据可以改善疾病基因的预测,并有助于进一步了解疾病的发病机理。我们使用交叉验证验证了我们的模型结构,并通过蛋白质关联网络分析验证了我们的优先排序结果,结果表明,与非优先排序的基因相比,优先排序的CD基因相互作用更高。尽管单个数据集无法传达与疾病相关的所有信息,但将多个基于表达的相关数据集相结合的数据可以改善疾病基因的预测,并有助于进一步了解疾病的发病机理。我们使用交叉验证验证了我们的模型结构,并通过蛋白质关联网络分析验证了我们的优先排序结果,结果表明,与非优先排序的基因相比,优先排序的CD基因相互作用更高。尽管单个数据集无法传达与疾病相关的所有信息,但将多个基于表达的相关数据集相结合的数据可以改善疾病基因的预测,并有助于进一步了解疾病的发病机理。


























 京公网安备 11010802027423号
京公网安备 11010802027423号