PLOS ONE ( IF 3.7 ) Pub Date : 2019-01-17 , DOI: 10.1371/journal.pone.0210970 Dennis Versluis 1 , Teresita de J Bello González 1 , Erwin G Zoetendal 1 , Mark W J van Passel 1, 2 , Hauke Smidt 1
|
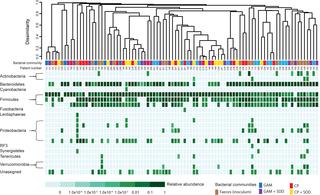
The emergence of bacterial pathogens that are resistant to clinical antibiotics poses an increasing risk to human health. An important reservoir from which bacterial pathogens can acquire resistance is the human gut microbiota. However, thus far, a substantial fraction of the gut microbiota remains uncultivated and has been little-studied with respect to its resistance reservoir-function. Here, we aimed to isolate yet uncultivated resistant gut bacteria by a targeted approach. Therefore, faecal samples from 20 intensive care patients who had received the prophylactic antibiotic treatment selective digestive decontamination (SDD), i.e. tobramycin, polymyxin E, amphotericin B and cefotaxime, were inoculated anaerobically on porous aluminium oxide chips placed on top of poor and rich agar media, including media supplemented with the SDD antibiotics. Biomass growing on the chips was analysed by 16S rRNA gene amplicon sequencing, showing large inter-individual differences in bacterial cultivability, and enrichment of a range of taxonomically diverse operational taxonomic units (OTUs). Furthermore, growth of Ruminococcaceae (2 OTUs), Enterobacteriaceae (6 OTUs) and Lachnospiraceae (4 OTUs) was significantly inhibited by the SDD antibiotics. Strains belonging to 16 OTUs were candidates for cultivation to pure culture as they shared ≤95% sequence identity with the closest type strain and had a relative abundance of ≥2%. Six of these OTUs were detected on media containing SDD antibiotics, and as such were prime candidates to be studied regarding antibiotic resistance. One of these six OTUs was obtained in pure culture using targeted isolation. This novel strain was resistant to the antibiotics metrodinazole and imipenem. It was initially classified as member of the Ruminococcaceae, though later it was found to share 99% nucleotide identity with the recently published Sellimonas intestinalis BR72T. In conclusion, we show that high-throughput cultivation-based screening of microbial communities can guide targeted isolation of bacteria that serve as reservoirs of antibiotic resistance.
中文翻译:

基于多孔氧化铝芯片的高通量培养筛选可以有针对性地分离抗生素耐药性人类肠道细菌
对临床抗生素具有抗药性的细菌病原体的出现对人类健康构成了越来越大的风险。细菌病原体获得抵抗力的一个重要储存库是人类肠道微生物群。然而,到目前为止,肠道微生物群的很大一部分仍未被培养,并且对其耐药库功能的研究很少。在这里,我们的目标是通过有针对性的方法分离尚未培养的耐药肠道细菌。因此,将来自20名接受预防性抗生素治疗选择性消化道净化(SDD)(即妥布霉素、多粘菌素E、两性霉素B和头孢噻肟)的重症监护患者的粪便样本厌氧接种到置于贫琼脂和富琼脂顶部的多孔氧化铝芯片上培养基,包括补充有 SDD 抗生素的培养基。通过 16S rRNA 基因扩增子测序对芯片上生长的生物质进行了分析,结果显示细菌可培养性存在巨大的个体间差异,并且富集了一系列分类学上多样化的操作分类单位 (OTU)。此外,SDD抗生素显着抑制瘤胃球菌科(2个OTU)、肠杆菌科(6个OTU)和毛螺菌科(4个OTU)的生长。属于 16 个 OTU 的菌株是纯培养的候选菌株,因为它们与最接近的类型菌株具有 ≤95% 的序列同一性,并且相对丰度≥2%。其中六个 OTU 在含有 SDD 抗生素的培养基上检测到,因此是研究抗生素耐药性的主要候选者。这六个 OTU 之一是使用靶向分离在纯培养物中获得的。这种新菌株对抗生素甲硝唑和亚胺培南具有耐药性。它最初被归类为瘤胃球菌科成员,但后来发现它与最近发表的肠小单胞菌BR72 T具有 99% 的核苷酸同一性。总之,我们表明,基于高通量培养的微生物群落筛选可以指导有针对性地分离作为抗生素耐药性库的细菌。



























 京公网安备 11010802027423号
京公网安备 11010802027423号