当前位置:
X-MOL 学术
›
ACS Cent. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
A Rapid and Low-Cost Pathogen Detection Platform by Using a Molecular Agglutination Assay
ACS Central Science ( IF 18.2 ) Pub Date : 2018-11-05 00:00:00 , DOI: 10.1021/acscentsci.8b00447 Tsung-Feng Wu 1 , Yu-Chen Chen 1 , Wei-Chung Wang 1 , Yen-Chi Fang 1 , Scott Fukuoka 2 , David T. Pride 3 , On Shun Pak 4
ACS Central Science ( IF 18.2 ) Pub Date : 2018-11-05 00:00:00 , DOI: 10.1021/acscentsci.8b00447 Tsung-Feng Wu 1 , Yu-Chen Chen 1 , Wei-Chung Wang 1 , Yen-Chi Fang 1 , Scott Fukuoka 2 , David T. Pride 3 , On Shun Pak 4
Affiliation
|
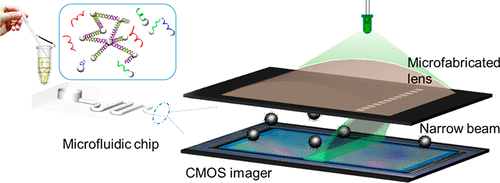
Rapid and low-cost pathogen diagnostic approaches are critical for clinical decision-making procedures. Cultivating bacteria often takes days to identify pathogens and provide antimicrobial susceptibilities. The delay in diagnosis may result in compromised treatment and inappropriate antibiotic use. Over the past decades, molecular-based techniques have significantly shortened pathogen identification turnaround time with high accuracy. However, these assays often use complex fluorescent labeling and nucleic acid amplification processes, which limit their use in resource-limited settings. In this work, we demonstrate a wash-free molecular agglutination assay with a straightforward mixing and incubation step that significantly simplifies procedures of molecular testing. By targeting the 16S rRNA gene of pathogens, we perform a rapid pathogen identification within 30 min on a dark-field imaging microfluidic cytometry platform. The dark-field images with low background noise can be obtained using a narrow beam scanning technique with off-the-shelf complementary metal oxide semiconductor (CMOS) imagers such as smartphone cameras. We utilize a machine learning algorithm to deconvolute topological features of agglutinated clusters and thus quantify the abundance of bacteria. Consequently, we unambiguously distinguish Escherichia coli positive from other E. coli negative among 50 clinical urinary tract infection samples with 96% sensitivity and 100% specificity. Furthermore, we also apply this quantitative detection approach to achieve rapid antimicrobial susceptibility testing within 3 h. This work exhibits easy-to-use protocols, high sensitivity, and short turnaround time for point-of-care testing uses.
中文翻译:

分子凝集法快速低成本的病原体检测平台
快速和低成本的病原体诊断方法对于临床决策程序至关重要。培养细菌通常需要几天的时间来鉴定病原体并提供抗菌药敏性。诊断的延迟可能会导致治疗受损和抗生素使用不当。在过去的几十年中,基于分子的技术以高精度大大缩短了病原体鉴定的周转时间。但是,这些测定通常使用复杂的荧光标记和核酸扩增过程,这限制了它们在资源有限的环境中的使用。在这项工作中,我们展示了一种免洗的分子凝集测定法,它具有简单的混合和孵育步骤,可大大简化分子测试的程序。通过靶向病原体的16S rRNA基因,我们在暗场成像微流式细胞仪平台上的30分钟内执行了快速的病原体鉴定。可以使用窄束扫描技术和现成的互补金属氧化物半导体(CMOS)成像器(如智能手机相机)使用窄束扫描技术获得具有低背景噪声的暗场图像。我们利用机器学习算法对聚集簇的拓扑特征进行反卷积,从而量化细菌的丰度。因此,我们可以明确地区分 我们利用机器学习算法对聚集簇的拓扑特征进行反卷积,从而量化细菌的丰度。因此,我们可以明确地区分 我们利用机器学习算法对聚集簇的拓扑特征进行反卷积,从而量化细菌的丰度。因此,我们可以明确地区分在50个临床尿道感染样本中,大肠杆菌与其他大肠杆菌阴性呈阳性,敏感性为96%,特异性为100%。此外,我们还应用这种定量检测方法在3小时内实现了快速的抗生素敏感性测试。这项工作表现出易于使用的协议,高灵敏度和较短的周转时间,可用于即时医疗测试。
更新日期:2018-11-05
中文翻译:

分子凝集法快速低成本的病原体检测平台
快速和低成本的病原体诊断方法对于临床决策程序至关重要。培养细菌通常需要几天的时间来鉴定病原体并提供抗菌药敏性。诊断的延迟可能会导致治疗受损和抗生素使用不当。在过去的几十年中,基于分子的技术以高精度大大缩短了病原体鉴定的周转时间。但是,这些测定通常使用复杂的荧光标记和核酸扩增过程,这限制了它们在资源有限的环境中的使用。在这项工作中,我们展示了一种免洗的分子凝集测定法,它具有简单的混合和孵育步骤,可大大简化分子测试的程序。通过靶向病原体的16S rRNA基因,我们在暗场成像微流式细胞仪平台上的30分钟内执行了快速的病原体鉴定。可以使用窄束扫描技术和现成的互补金属氧化物半导体(CMOS)成像器(如智能手机相机)使用窄束扫描技术获得具有低背景噪声的暗场图像。我们利用机器学习算法对聚集簇的拓扑特征进行反卷积,从而量化细菌的丰度。因此,我们可以明确地区分 我们利用机器学习算法对聚集簇的拓扑特征进行反卷积,从而量化细菌的丰度。因此,我们可以明确地区分 我们利用机器学习算法对聚集簇的拓扑特征进行反卷积,从而量化细菌的丰度。因此,我们可以明确地区分在50个临床尿道感染样本中,大肠杆菌与其他大肠杆菌阴性呈阳性,敏感性为96%,特异性为100%。此外,我们还应用这种定量检测方法在3小时内实现了快速的抗生素敏感性测试。这项工作表现出易于使用的协议,高灵敏度和较短的周转时间,可用于即时医疗测试。


























 京公网安备 11010802027423号
京公网安备 11010802027423号