当前位置:
X-MOL 学术
›
ACS Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Genetic Code Expansion Method for Temporal Labeling of Endogenously Expressed Proteins
ACS Chemical Biology ( IF 4 ) Pub Date : 2018-10-10 00:00:00 , DOI: 10.1021/acschembio.8b00594 Nils Schneider 1, 2 , Christoph Gäbelein 1, 2 , Julius Wiener 1, 2 , Tihomir Georgiev 3 , Nicolas Gobet 1, 2 , Wilfried Weber 2, 4 , Matthias Meier 1, 2, 3
ACS Chemical Biology ( IF 4 ) Pub Date : 2018-10-10 00:00:00 , DOI: 10.1021/acschembio.8b00594 Nils Schneider 1, 2 , Christoph Gäbelein 1, 2 , Julius Wiener 1, 2 , Tihomir Georgiev 3 , Nicolas Gobet 1, 2 , Wilfried Weber 2, 4 , Matthias Meier 1, 2, 3
Affiliation
|
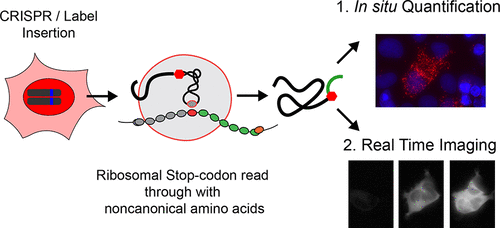
We here present a method that combines genetic code expansion with CRISPR/Cas9 genome engineering to label endogenously expressed proteins with high spatiotemporal resolution. The method exploits the use of an orthogonal tRNA/tRNA synthetase pair in conjugation with noncanonical amino acids to create stop codon read through events. To demonstrate the functionality of the method, we pulse labeled endogenous β-actin and tumor protein p53 with a minimally invasive HA tag at their C-termini. Targeting the protein label with a proximity ligation assay plus real time imaging facilitates seamless quantification of the protein synthesis rate and spatial localization at the single cell level. The presented approach does not interfere with any physiological control of cellular expression, nor did we observe any perturbation of endogenous protein functions.
中文翻译:

内源表达蛋白的时间标记的遗传密码扩展方法
我们在这里提出了一种将遗传密码扩展与CRISPR / Cas9基因组工程相结合的方法,以高时空分辨率标记内源表达的蛋白质。该方法利用了与非经典氨基酸结合使用正交tRNA / tRNA合成酶对来创建终止密码子通读事件。为了证明该方法的功能,我们用标记的内源性β-肌动蛋白和肿瘤蛋白p53在其C末端加上了微创HA标签。通过邻近连接测定法和实时成像来靶向蛋白质标签,有助于在单个细胞水平上对蛋白质合成速率和空间定位进行无缝定量。提出的方法不会干扰细胞表达的任何生理控制,我们也没有观察到任何内源蛋白功能的扰动。
更新日期:2018-10-10
中文翻译:

内源表达蛋白的时间标记的遗传密码扩展方法
我们在这里提出了一种将遗传密码扩展与CRISPR / Cas9基因组工程相结合的方法,以高时空分辨率标记内源表达的蛋白质。该方法利用了与非经典氨基酸结合使用正交tRNA / tRNA合成酶对来创建终止密码子通读事件。为了证明该方法的功能,我们用标记的内源性β-肌动蛋白和肿瘤蛋白p53在其C末端加上了微创HA标签。通过邻近连接测定法和实时成像来靶向蛋白质标签,有助于在单个细胞水平上对蛋白质合成速率和空间定位进行无缝定量。提出的方法不会干扰细胞表达的任何生理控制,我们也没有观察到任何内源蛋白功能的扰动。


























 京公网安备 11010802027423号
京公网安备 11010802027423号