当前位置:
X-MOL 学术
›
Mol. Syst. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Multi‐Omics Factor Analysis—a framework for unsupervised integration of multi‐omics data sets
Molecular Systems Biology ( IF 9.9 ) Pub Date : 2018-06-20 , DOI: 10.15252/msb.20178124 Ricard Argelaguet 1 , Britta Velten 2 , Damien Arnol 1 , Sascha Dietrich 3 , Thorsten Zenz 3, 4, 5 , John C Marioni 1, 6, 7 , Florian Buettner 8, 9 , Wolfgang Huber 10 , Oliver Stegle 2, 8
Molecular Systems Biology ( IF 9.9 ) Pub Date : 2018-06-20 , DOI: 10.15252/msb.20178124 Ricard Argelaguet 1 , Britta Velten 2 , Damien Arnol 1 , Sascha Dietrich 3 , Thorsten Zenz 3, 4, 5 , John C Marioni 1, 6, 7 , Florian Buettner 8, 9 , Wolfgang Huber 10 , Oliver Stegle 2, 8
Affiliation
|
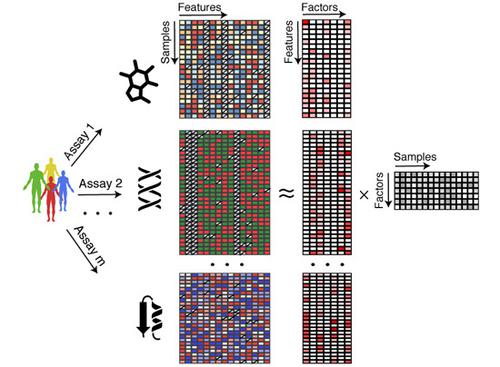
Multi‐omics studies promise the improved characterization of biological processes across molecular layers. However, methods for the unsupervised integration of the resulting heterogeneous data sets are lacking. We present Multi‐Omics Factor Analysis (MOFA), a computational method for discovering the principal sources of variation in multi‐omics data sets. MOFA infers a set of (hidden) factors that capture biological and technical sources of variability. It disentangles axes of heterogeneity that are shared across multiple modalities and those specific to individual data modalities. The learnt factors enable a variety of downstream analyses, including identification of sample subgroups, data imputation and the detection of outlier samples. We applied MOFA to a cohort of 200 patient samples of chronic lymphocytic leukaemia, profiled for somatic mutations, RNA expression, DNA methylation and ex vivo drug responses. MOFA identified major dimensions of disease heterogeneity, including immunoglobulin heavy‐chain variable region status, trisomy of chromosome 12 and previously underappreciated drivers, such as response to oxidative stress. In a second application, we used MOFA to analyse single‐cell multi‐omics data, identifying coordinated transcriptional and epigenetic changes along cell differentiation.
中文翻译:

多组学因子分析——多组学数据集的无监督集成框架
多组学研究有望改进跨分子层的生物过程表征。然而,缺乏对生成的异构数据集进行无监督集成的方法。我们提出了多组学因子分析 (MOFA),这是一种用于发现多组学数据集中主要变异来源的计算方法。MOFA 推断出一组(隐藏的)因素,这些因素可以捕捉到变异的生物和技术来源。它解开了跨多种模式共享的异质性轴和特定于单个数据模式的轴。学习到的因素可以进行各种下游分析,包括样本子组的识别、数据插补和异常样本的检测。我们将 MOFA 应用于一组 200 名慢性淋巴细胞白血病患者样本,离体药物反应。MOFA 确定了疾病异质性的主要方面,包括免疫球蛋白重链可变区状态、12 号染色体三体性和以前被低估的驱动因素,例如对氧化应激的反应。在第二个应用中,我们使用 MOFA 分析单细胞多组学数据,识别细胞分化过程中协调的转录和表观遗传变化。
更新日期:2019-11-18
中文翻译:

多组学因子分析——多组学数据集的无监督集成框架
多组学研究有望改进跨分子层的生物过程表征。然而,缺乏对生成的异构数据集进行无监督集成的方法。我们提出了多组学因子分析 (MOFA),这是一种用于发现多组学数据集中主要变异来源的计算方法。MOFA 推断出一组(隐藏的)因素,这些因素可以捕捉到变异的生物和技术来源。它解开了跨多种模式共享的异质性轴和特定于单个数据模式的轴。学习到的因素可以进行各种下游分析,包括样本子组的识别、数据插补和异常样本的检测。我们将 MOFA 应用于一组 200 名慢性淋巴细胞白血病患者样本,离体药物反应。MOFA 确定了疾病异质性的主要方面,包括免疫球蛋白重链可变区状态、12 号染色体三体性和以前被低估的驱动因素,例如对氧化应激的反应。在第二个应用中,我们使用 MOFA 分析单细胞多组学数据,识别细胞分化过程中协调的转录和表观遗传变化。



























 京公网安备 11010802027423号
京公网安备 11010802027423号