当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Ligand-Guided Selection with Artificially Expanded Genetic Information Systems against TCR-CD3ε.
Biochemistry ( IF 2.9 ) Pub Date : 2020-01-08 , DOI: 10.1021/acs.biochem.9b00919 Hasan Zumrut 1 , Zunyi Yang 2 , Nicole Williams 3 , Joekeem Arizala 1 , Sana Batool 4 , Steven A Benner 2, 5 , Prabodhika Mallikaratchy 1, 3, 4
Biochemistry ( IF 2.9 ) Pub Date : 2020-01-08 , DOI: 10.1021/acs.biochem.9b00919 Hasan Zumrut 1 , Zunyi Yang 2 , Nicole Williams 3 , Joekeem Arizala 1 , Sana Batool 4 , Steven A Benner 2, 5 , Prabodhika Mallikaratchy 1, 3, 4
Affiliation
|
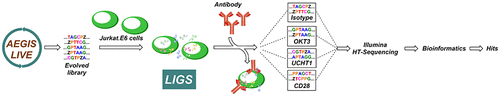
Here we are reporting, for the first time, a ligand-guided selection (LIGS) experiment using an artificially expanded genetic information system (AEGIS) to successfully identify an AEGIS-DNA aptamer against T cell receptor-CD3ε expressed on Jurkat.E6 cells. Thus, we have effectively combined the enhanced diversity of an AEGIS DNA library with LIGS to develop a superior screening platform to discover superior aptamers. Libraries of DNA molecules from highly diversified building blocks will provide better ligands due to more functional diversity and better-controlled folding. Thus, a DNA library with AEGIS components (dZ and dP) was used in LIGS experiments against TCR-CD3ε in its native state using two clinically relevant monoclonal antibodies to identify an aptamer termed JZPO-10, with nanomolar affinity. Multiple specificity assays using knockout cells, and competition experiments using monoclonal antibodies utilized in LIGS, show unprecedented specificity of JZPO-10, suggesting that the combination of LIGS with AEGIS-DNA libraries will provide a superior screening platform to discover artificial ligands against critical cellular targets.
中文翻译:

针对TCR-CD3ε的人工扩展遗传信息系统的配体引导选择。
在这里,我们首次报道了使用人工扩展的遗传信息系统(AEGIS)进行的配体引导选择(LIGS)实验,以成功识别针对Jurkat.E6细胞上表达的T细胞受体CD3ε的AEGIS-DNA适体。因此,我们有效地将AEGIS DNA库的增强多样性与LIGS结合在一起,以开发出卓越的筛选平台来发现卓越的适体。高度多样化的构建基团的DNA分子文库将提供更好的配体,因为它们具有更多的功能多样性和更好的控制折叠性。因此,具有AEGIS成分(dZ和dP)的DNA文库用于LIGS实验中,针对天然状态的TCR-CD3ε,使用两种临床相关的单克隆抗体来鉴定具有纳摩尔摩尔亲和力的称为JZPO-10的适体。
更新日期:2020-01-08
中文翻译:

针对TCR-CD3ε的人工扩展遗传信息系统的配体引导选择。
在这里,我们首次报道了使用人工扩展的遗传信息系统(AEGIS)进行的配体引导选择(LIGS)实验,以成功识别针对Jurkat.E6细胞上表达的T细胞受体CD3ε的AEGIS-DNA适体。因此,我们有效地将AEGIS DNA库的增强多样性与LIGS结合在一起,以开发出卓越的筛选平台来发现卓越的适体。高度多样化的构建基团的DNA分子文库将提供更好的配体,因为它们具有更多的功能多样性和更好的控制折叠性。因此,具有AEGIS成分(dZ和dP)的DNA文库用于LIGS实验中,针对天然状态的TCR-CD3ε,使用两种临床相关的单克隆抗体来鉴定具有纳摩尔摩尔亲和力的称为JZPO-10的适体。



























 京公网安备 11010802027423号
京公网安备 11010802027423号