当前位置:
X-MOL 学术
›
Eur. J. Pharm. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Discovery of potential 1,3,5-Triazine compounds against strains of Plasmodium falciparum using supervised machine learning models.
European Journal of Pharmaceutical Sciences ( IF 4.6 ) Pub Date : 2019-12-25 , DOI: 10.1016/j.ejps.2019.105208 Supriya Sahu 1 , Surajit Kumar Ghosh 1 , Jun Moni Kalita 1 , Murali C Ginjupalli 2 , Kranthi Raj K 3
European Journal of Pharmaceutical Sciences ( IF 4.6 ) Pub Date : 2019-12-25 , DOI: 10.1016/j.ejps.2019.105208 Supriya Sahu 1 , Surajit Kumar Ghosh 1 , Jun Moni Kalita 1 , Murali C Ginjupalli 2 , Kranthi Raj K 3
Affiliation
|
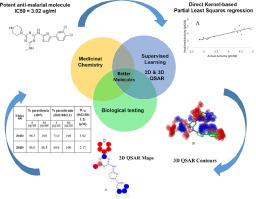
The Malaria burden was an escalating global encumbrance and need to be addressed with critical care. Anti-malarial drug discovery was integrated with supervised machine learning (ML) models to identify potent thiazolyl-traizine derivatives. This assimilated approach of Direct Kernel-based Partial Least Squares regression (DKPLS) with molprint 2D fingerprints in Quantitative Structure Activity Relationship models was utilized to map the knowledge of known actives and to design novel molecules. This QSAR study had revealed the structural features required for better antimalarial activity. Two of the molecules which were designed based on the results of this QSAR study, had shown good percentage of parasitemia against both chloroquine sensitive (3D7) and chloroquine resistant (Dd2) strains of Plasmodium falciparum respectively. The IC50 of 201D and 204D was 3.02 and 2.17 µM against chloroquine resistant Dd2 strain of Plasmodium falciparum. This result had proved the efficiency of a multidisciplinary approach of medicinal chemistry and machine learning for the design of novel potent anti-malarial compounds.
中文翻译:

使用监督的机器学习模型发现针对恶性疟原虫菌株的潜在1,3,5-三嗪化合物。
疟疾负担是全球范围内不断升级的负担,需要给予重症监护。将抗疟疾药物发现与有监督的机器学习(ML)模型集成在一起,以鉴定有效的噻唑基-traizine衍生物。在定量结构活性关系模型中,这种基于直接内核的偏最小二乘回归(DKPLS)与molprint 2D指纹的同化方法被用来绘制已知活性成分的知识并设计新分子。这项QSAR研究揭示了更好的抗疟疾活性所需的结构特征。基于该QSAR研究结果设计的两个分子分别显示出针对恶性疟原虫的氯喹敏感(3D7)和耐氯喹(Dd2)菌株的高寄生率。201D和204D对恶性疟原虫抗氯喹Dd2菌株的IC50为3.02和2.17 µM。该结果证明了药物化学和机器学习的多学科方法对于新型有效的抗疟疾化合物的设计是有效的。
更新日期:2019-12-26
中文翻译:

使用监督的机器学习模型发现针对恶性疟原虫菌株的潜在1,3,5-三嗪化合物。
疟疾负担是全球范围内不断升级的负担,需要给予重症监护。将抗疟疾药物发现与有监督的机器学习(ML)模型集成在一起,以鉴定有效的噻唑基-traizine衍生物。在定量结构活性关系模型中,这种基于直接内核的偏最小二乘回归(DKPLS)与molprint 2D指纹的同化方法被用来绘制已知活性成分的知识并设计新分子。这项QSAR研究揭示了更好的抗疟疾活性所需的结构特征。基于该QSAR研究结果设计的两个分子分别显示出针对恶性疟原虫的氯喹敏感(3D7)和耐氯喹(Dd2)菌株的高寄生率。201D和204D对恶性疟原虫抗氯喹Dd2菌株的IC50为3.02和2.17 µM。该结果证明了药物化学和机器学习的多学科方法对于新型有效的抗疟疾化合物的设计是有效的。


























 京公网安备 11010802027423号
京公网安备 11010802027423号