当前位置:
X-MOL 学术
›
Phys. Chem. Chem. Phys.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Calculation of salt-dependent free energy of binding of β-lactoglobulin homodimer formation and mechanism of dimer formation using molecular dynamics simulation and three-dimensional reference interaction site model (3D-RISM): diffuse salt ions and non-polar interactions between the monomers favor the dimer formation.
Physical Chemistry Chemical Physics ( IF 3.3 ) Pub Date : 2020-01-08 , DOI: 10.1039/c9cp05578a Rakesh Srivastava 1 , Mausumi Chattopadhyaya 2 , Pradipta Bandyopadhyay 1
Physical Chemistry Chemical Physics ( IF 3.3 ) Pub Date : 2020-01-08 , DOI: 10.1039/c9cp05578a Rakesh Srivastava 1 , Mausumi Chattopadhyaya 2 , Pradipta Bandyopadhyay 1
Affiliation
|
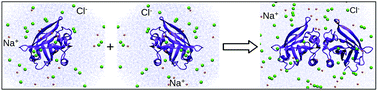
There are several important phenomena in chemistry, biology, and physics where molecules (or parts of a molecule) having charges of the same sign come closer together and become stable. DNA condensation, RNA folding, colloid-colloid interactions are some of the examples of this kind. In the current work, we have investigated how β-lactoglobulin, a protein found in milk, in spite of carrying +13 charge, favors the homodimer formation in the presence of salt. We have focussed on calculating the protein-protein binding free energy in the presence of salt and identifying the thermodynamic and microscopic mechanism of the process. Estimation of binding free energy of this salt-dependent process is done by combining molecular dynamics simulation with statistical mechanical theory of three-dimensional reference interaction site model (3D-RISM). Binding free energy is evaluated from the chemical potential of the solutes as opposed to potential of mean force calculation, which gives only a constrained free energy. Our calculated values semi-quantitatively match with the experimental results. By examining the different components of binding free energy, we have found that the role of salt ions (especially of Cl-) is to shift the equilibrium towards the dimer. Non-polar (Lennard-Jones) interactions between the monomers is also favorable to the binding free energy. However, water slightly disfavors the dimer formation. For the microscopic mechanism, heterogeneous of both Na+ and Cl- near the charged residues at the binding interface and change of this charge distribution on dimer formation contribute to the stability. A fine-tuning of enthalpic and entropic effects of salt ions is found to operate at different salt concentrations. Both thermodynamic and microscopic mechanism of dimer formation gives detailed insight into the complex electrostatics of charged protein-protein binding.
中文翻译:

使用分子动力学模拟和三维参考相互作用位点模型(3D-RISM)计算β-乳球蛋白同二聚体结合的盐依赖性自由能和二聚体形成机理:分散的盐离子和单体之间的非极性相互作用二聚体形成。
在化学,生物学和物理学中,有几个重要的现象,其中具有相同符号的分子(或分子的一部分)彼此靠近并变得稳定。DNA缩合,RNA折叠,胶体-胶体相互作用就是这类例子。在当前的工作中,我们研究了牛奶中发现的一种蛋白质β-乳球蛋白,尽管带有+13电荷,但在有盐的情况下如何促进同型二聚体的形成。我们专注于在存在盐的情况下计算蛋白质与蛋白质的结合自由能,并确定该过程的热力学和微观机理。通过将分子动力学模拟与三维参考相互作用位点模型(3D-RISM)的统计力学理论相结合,可以估算此盐依赖性过程的结合自由能。结合自由能是从溶质的化学势来评估的,而与平均力计算的势相反,后者仅给出了受约束的自由能。我们的计算值与实验结果半定量匹配。通过检查结合自由能的不同成分,我们发现盐离子(尤其是Cl-)的作用是使平衡向二聚体移动。单体之间的非极性(兰纳德-琼斯)相互作用也有利于结合自由能。但是,水稍微不利于二聚体的形成。对于微观机理,在结合界面上靠近带电残基的Na +和Cl-均不均匀,并且这种电荷分布在二聚体形成上的变化有助于稳定性。发现盐离子的焓和熵效应的微调在不同的盐浓度下起作用。二聚体形成的热力学和微观机理都提供了对带电蛋白质-蛋白质结合的复杂静电的详细了解。
更新日期:2020-01-08
中文翻译:

使用分子动力学模拟和三维参考相互作用位点模型(3D-RISM)计算β-乳球蛋白同二聚体结合的盐依赖性自由能和二聚体形成机理:分散的盐离子和单体之间的非极性相互作用二聚体形成。
在化学,生物学和物理学中,有几个重要的现象,其中具有相同符号的分子(或分子的一部分)彼此靠近并变得稳定。DNA缩合,RNA折叠,胶体-胶体相互作用就是这类例子。在当前的工作中,我们研究了牛奶中发现的一种蛋白质β-乳球蛋白,尽管带有+13电荷,但在有盐的情况下如何促进同型二聚体的形成。我们专注于在存在盐的情况下计算蛋白质与蛋白质的结合自由能,并确定该过程的热力学和微观机理。通过将分子动力学模拟与三维参考相互作用位点模型(3D-RISM)的统计力学理论相结合,可以估算此盐依赖性过程的结合自由能。结合自由能是从溶质的化学势来评估的,而与平均力计算的势相反,后者仅给出了受约束的自由能。我们的计算值与实验结果半定量匹配。通过检查结合自由能的不同成分,我们发现盐离子(尤其是Cl-)的作用是使平衡向二聚体移动。单体之间的非极性(兰纳德-琼斯)相互作用也有利于结合自由能。但是,水稍微不利于二聚体的形成。对于微观机理,在结合界面上靠近带电残基的Na +和Cl-均不均匀,并且这种电荷分布在二聚体形成上的变化有助于稳定性。发现盐离子的焓和熵效应的微调在不同的盐浓度下起作用。二聚体形成的热力学和微观机理都提供了对带电蛋白质-蛋白质结合的复杂静电的详细了解。



























 京公网安备 11010802027423号
京公网安备 11010802027423号