当前位置:
X-MOL 学术
›
Nat. Genet.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Identifying cross-disease components of genetic risk across hospital data in the UK Biobank.
Nature Genetics ( IF 30.8 ) Pub Date : 2019-12-23 , DOI: 10.1038/s41588-019-0550-4 Adrian Cortes 1, 2 , Patrick K Albers 1 , Calliope A Dendrou 3 , Lars Fugger 2, 4, 5 , Gil McVean 1
Nature Genetics ( IF 30.8 ) Pub Date : 2019-12-23 , DOI: 10.1038/s41588-019-0550-4 Adrian Cortes 1, 2 , Patrick K Albers 1 , Calliope A Dendrou 3 , Lars Fugger 2, 4, 5 , Gil McVean 1
Affiliation
|
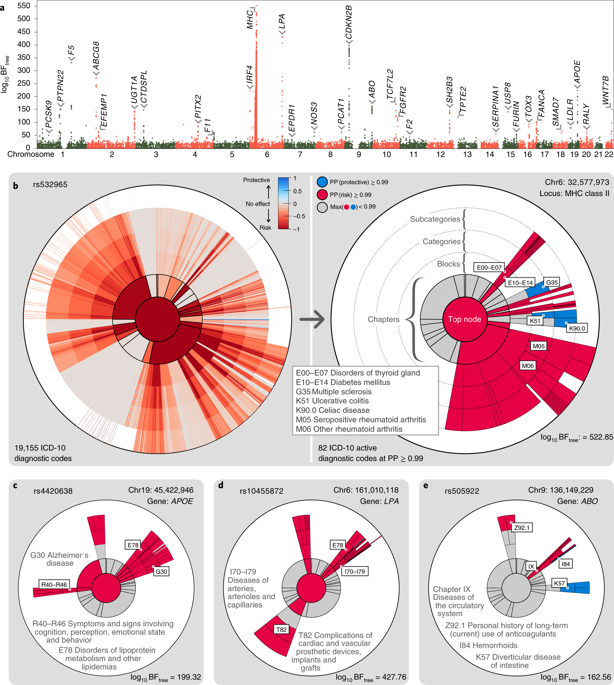
Genetic risk factors frequently affect multiple common human diseases, providing insight into shared pathophysiological pathways and opportunities for therapeutic development. However, systematic identification of genetic profiles of disease risk is limited by the availability of both comprehensive clinical data on population-scale cohorts and the lack of suitable statistical methodology that can handle the scale of and differential power inherent in multi-phenotype data. Here, we develop a disease-agnostic approach to cluster the genetic risk profiles for 3,025 genome-wide independent loci across 19,155 disease classification codes from 320,644 participants in the UK Biobank, representing a large and heterogeneous population. We identify 339 distinct disease association profiles and use multiple approaches to link clusters to the underlying biological pathways. We show how clusters can decompose the variance and covariance in risk for disease, thereby identifying underlying biological processes and their impact. We demonstrate the use of clusters in defining disease relationships and their potential in informing therapeutic strategies.
中文翻译:

在英国生物银行的医院数据中识别遗传风险的跨疾病成分。
遗传风险因素经常影响多种常见的人类疾病,提供对共同的病理生理途径和治疗发展机会的洞察。然而,疾病风险遗传谱的系统识别受到人口规模队列综合临床数据的可用性以及缺乏合适的统计方法来处理多表型数据中固有的规模和差异能力的限制。在这里,我们开发了一种与疾病无关的方法,对来自英国生物库的 320,644 名参与者的 19,155 个疾病分类代码中的 3,025 个全基因组独立位点的遗传风险谱进行聚类,这些参与者代表了一个庞大且异质的人群。我们确定了 339 种不同的疾病关联概况,并使用多种方法将集群与潜在的生物途径联系起来。我们展示了集群如何分解疾病风险的方差和协方差,从而识别潜在的生物过程及其影响。我们展示了集群在定义疾病关系中的用途以及它们在为治疗策略提供信息方面的潜力。
更新日期:2019-12-23
中文翻译:

在英国生物银行的医院数据中识别遗传风险的跨疾病成分。
遗传风险因素经常影响多种常见的人类疾病,提供对共同的病理生理途径和治疗发展机会的洞察。然而,疾病风险遗传谱的系统识别受到人口规模队列综合临床数据的可用性以及缺乏合适的统计方法来处理多表型数据中固有的规模和差异能力的限制。在这里,我们开发了一种与疾病无关的方法,对来自英国生物库的 320,644 名参与者的 19,155 个疾病分类代码中的 3,025 个全基因组独立位点的遗传风险谱进行聚类,这些参与者代表了一个庞大且异质的人群。我们确定了 339 种不同的疾病关联概况,并使用多种方法将集群与潜在的生物途径联系起来。我们展示了集群如何分解疾病风险的方差和协方差,从而识别潜在的生物过程及其影响。我们展示了集群在定义疾病关系中的用途以及它们在为治疗策略提供信息方面的潜力。


























 京公网安备 11010802027423号
京公网安备 11010802027423号