当前位置:
X-MOL 学术
›
J. Phys. Chem. B
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Structural Analysis of 14-3-3-ζ-Derived Phosphopeptides Using Electron Capture Dissociation Mass Spectrometry, Traveling Wave Ion Mobility Spectrometry, and Molecular Modeling.
The Journal of Physical Chemistry B ( IF 3.3 ) Pub Date : 2019-12-20 , DOI: 10.1021/acs.jpcb.9b08506 Anna L Simmonds , Andrea F Lopez-Clavijo , Peter J Winn , David H Russell 1 , Iain B Styles 2 , Helen J Cooper
The Journal of Physical Chemistry B ( IF 3.3 ) Pub Date : 2019-12-20 , DOI: 10.1021/acs.jpcb.9b08506 Anna L Simmonds , Andrea F Lopez-Clavijo , Peter J Winn , David H Russell 1 , Iain B Styles 2 , Helen J Cooper
Affiliation
|
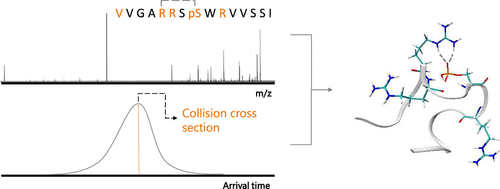
Previously, we have demonstrated the effect of salt bridges on the electron capture dissociation mass spectrometry behavior of synthetic model phosphopeptides and applied an ion mobility spectrometry/molecular modeling approach to rationalize the findings in terms of peptide ion structure. Here, we develop and apply the approach to a biologically derived phosphopeptide. Specifically, we have investigated variants of a 15-mer phosphopeptide VVGARRSsWRVVSSI (s denotes phosphorylated Ser) derived from Akt1 substrate 14-3-3-ζ, which contains the phosphorylation motif RRSsWR. Variants were generated by successive arginine-to-leucine substitutions within the phosphorylation motif. ECD fragmentation patterns for the eight phosphopeptide variants show greater sequence coverage with successive R → L substitutions. Peptides with two or more basic residues had regions with no sequence coverage, while full sequence coverage was observed for peptides with one or no basic residues. For three of the peptide variants, low-abundance fragments were observed between the phosphoserine and a basic residue, possibly due to the presence of multiple conformers with and without noncovalent interactions between these residues. For the five variants whose dissociation behavior suggested the presence of intramolecular noncovalent interactions, we employed ion mobility spectrometry and molecular modeling to probe the nature of these interactions. Our workflow allowed us to propose candidate structures whose noncovalent interactions were consistent with the ECD data for all of the peptides modeled. Additionally, the AMBER parameter sets created for and validated by this work are presented and made available online ( http://www.biosciences-labs.bham.ac.uk/cooper/datasets.php ).
中文翻译:

使用电子捕获解离质谱,行波离子迁移谱和分子建模对14-3-3--3-ζ衍生的磷酸肽进行结构分析。
以前,我们已经证明了盐桥对合成模型磷酸肽的电子捕获解离质谱行为的影响,并应用了离子迁移谱/分子建模方法来合理化肽离子结构方面的发现。在这里,我们开发并将该方法应用于生物衍生的磷酸肽。具体而言,我们研究了源自Akt1底物14-3-3-ζ的15-mer磷酸肽VVGARRSsWRVVSSI(s表示磷酸化Ser)的变体,其包含磷酸化基序RRSsWR。通过在磷酸化基序内连续的精氨酸至亮氨酸取代产生变体。八个磷酸肽变体的ECD片段化模式显示了更大的序列覆盖范围,具有连续的R→L取代。具有两个或多个碱性残基的肽具有无序列覆盖的区域,而对于具有一个或无碱性残基的肽则观察到全序列覆盖。对于三个肽变体,在磷酸丝氨酸和碱性残基之间观察到低丰度片段,可能是由于存在多个构象异构体,在这些残基之间存在非共价相互作用。对于五个解离行为表明存在分子内非共价相互作用的变体,我们采用了离子迁移谱和分子模型来探讨这些相互作用的性质。我们的工作流程使我们能够提出候选结构,该结构的非共价相互作用与所有建模肽的ECD数据均一致。此外,
更新日期:2020-01-10
中文翻译:

使用电子捕获解离质谱,行波离子迁移谱和分子建模对14-3-3--3-ζ衍生的磷酸肽进行结构分析。
以前,我们已经证明了盐桥对合成模型磷酸肽的电子捕获解离质谱行为的影响,并应用了离子迁移谱/分子建模方法来合理化肽离子结构方面的发现。在这里,我们开发并将该方法应用于生物衍生的磷酸肽。具体而言,我们研究了源自Akt1底物14-3-3-ζ的15-mer磷酸肽VVGARRSsWRVVSSI(s表示磷酸化Ser)的变体,其包含磷酸化基序RRSsWR。通过在磷酸化基序内连续的精氨酸至亮氨酸取代产生变体。八个磷酸肽变体的ECD片段化模式显示了更大的序列覆盖范围,具有连续的R→L取代。具有两个或多个碱性残基的肽具有无序列覆盖的区域,而对于具有一个或无碱性残基的肽则观察到全序列覆盖。对于三个肽变体,在磷酸丝氨酸和碱性残基之间观察到低丰度片段,可能是由于存在多个构象异构体,在这些残基之间存在非共价相互作用。对于五个解离行为表明存在分子内非共价相互作用的变体,我们采用了离子迁移谱和分子模型来探讨这些相互作用的性质。我们的工作流程使我们能够提出候选结构,该结构的非共价相互作用与所有建模肽的ECD数据均一致。此外,


























 京公网安备 11010802027423号
京公网安备 11010802027423号