当前位置:
X-MOL 学术
›
Nat. Rev. Genet.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Methods for mapping 3D chromosome architecture.
Nature Reviews Genetics ( IF 42.7 ) Pub Date : 2019-12-17 , DOI: 10.1038/s41576-019-0195-2 Rieke Kempfer 1, 2 , Ana Pombo 1, 2
Nature Reviews Genetics ( IF 42.7 ) Pub Date : 2019-12-17 , DOI: 10.1038/s41576-019-0195-2 Rieke Kempfer 1, 2 , Ana Pombo 1, 2
Affiliation
|
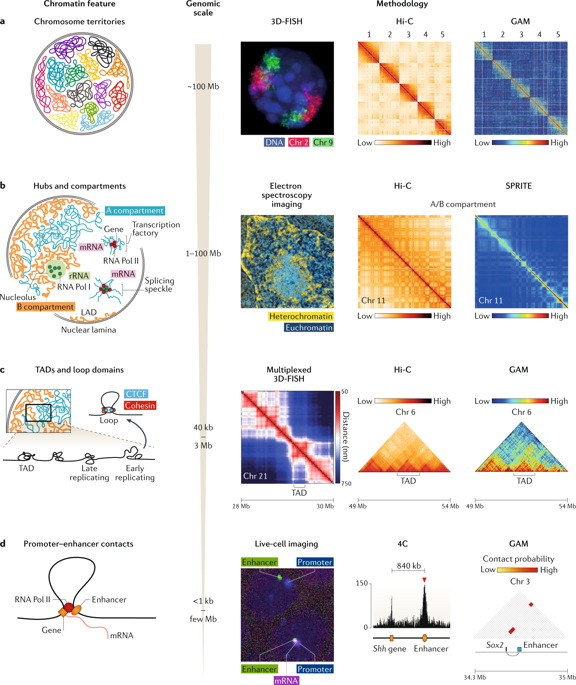
Determining how chromosomes are positioned and folded within the nucleus is critical to understanding the role of chromatin topology in gene regulation. Several methods are available for studying chromosome architecture, each with different strengths and limitations. Established imaging approaches and proximity ligation-based chromosome conformation capture (3C) techniques (such as DNA-FISH and Hi-C, respectively) have revealed the existence of chromosome territories, functional nuclear landmarks (such as splicing speckles and the nuclear lamina) and topologically associating domains. Improvements to these methods and the recent development of ligation-free approaches, including GAM, SPRITE and ChIA-Drop, are now helping to uncover new aspects of 3D genome topology that confirm the nucleus to be a complex, highly organized organelle.
中文翻译:

映射3D染色体体系结构的方法。
确定染色体如何在核内定位和折叠对于理解染色质拓扑在基因调控中的作用至关重要。有几种方法可用于研究染色体结构,每种方法具有不同的优势和局限性。既定的成像方法和基于邻近结扎的染色体构象捕获(3C)技术(分别为DNA-FISH和Hi-C)已揭示了染色体区域,功能性核标记(例如,拼接斑点和核层)的存在以及拓扑关联的域。这些方法的改进以及包括GAM,SPRITE和ChIA-Drop在内的无连接方法的最新发展现在正在帮助揭示3D基因组拓扑的新方面,这些新方面确认了核是一个复杂的,高度组织的细胞器。
更新日期:2019-12-17
中文翻译:

映射3D染色体体系结构的方法。
确定染色体如何在核内定位和折叠对于理解染色质拓扑在基因调控中的作用至关重要。有几种方法可用于研究染色体结构,每种方法具有不同的优势和局限性。既定的成像方法和基于邻近结扎的染色体构象捕获(3C)技术(分别为DNA-FISH和Hi-C)已揭示了染色体区域,功能性核标记(例如,拼接斑点和核层)的存在以及拓扑关联的域。这些方法的改进以及包括GAM,SPRITE和ChIA-Drop在内的无连接方法的最新发展现在正在帮助揭示3D基因组拓扑的新方面,这些新方面确认了核是一个复杂的,高度组织的细胞器。


























 京公网安备 11010802027423号
京公网安备 11010802027423号