当前位置:
X-MOL 学术
›
Blood Cancer J.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Comprehensive detection of recurring genomic abnormalities: a targeted sequencing approach for multiple myeloma.
Blood Cancer Journal ( IF 12.8 ) Pub Date : 2019-12-11 , DOI: 10.1038/s41408-019-0264-y Venkata Yellapantula 1 , Malin Hultcrantz 1, 2 , Even H Rustad 1 , Ester Wasserman 3 , Dory Londono 3 , Robert Cimera 3 , Amanda Ciardiello 1 , Heather Landau 4 , Theresia Akhlaghi 1 , Sham Mailankody 1 , Minal Patel 5 , Juan Santiago Medina-Martinez 5 , Juan Esteban Arango Ossa 5 , Max Fine Levine 5 , Niccolo Bolli 6, 7 , Francesco Maura 1 , Ahmet Dogan 8 , Elli Papaemmanuil 5, 9 , Yanming Zhang 3 , Ola Landgren 1
Blood Cancer Journal ( IF 12.8 ) Pub Date : 2019-12-11 , DOI: 10.1038/s41408-019-0264-y Venkata Yellapantula 1 , Malin Hultcrantz 1, 2 , Even H Rustad 1 , Ester Wasserman 3 , Dory Londono 3 , Robert Cimera 3 , Amanda Ciardiello 1 , Heather Landau 4 , Theresia Akhlaghi 1 , Sham Mailankody 1 , Minal Patel 5 , Juan Santiago Medina-Martinez 5 , Juan Esteban Arango Ossa 5 , Max Fine Levine 5 , Niccolo Bolli 6, 7 , Francesco Maura 1 , Ahmet Dogan 8 , Elli Papaemmanuil 5, 9 , Yanming Zhang 3 , Ola Landgren 1
Affiliation
|
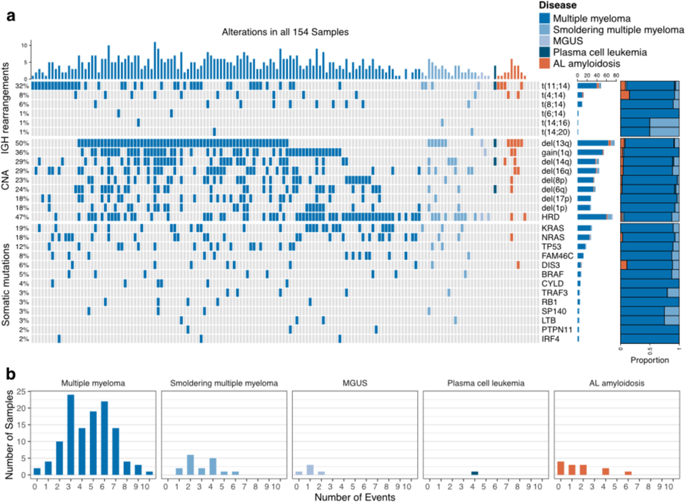
Recent genomic research efforts in multiple myeloma have revealed clinically relevant molecular subgroups beyond conventional cytogenetic classifications. Implementing these advances in clinical trial design and in routine patient care requires a new generation of molecular diagnostic tools. Here, we present a custom capture next-generation sequencing (NGS) panel designed to identify rearrangements involving the IGH locus, arm level, and focal copy number aberrations, as well as frequently mutated genes in multiple myeloma in a single assay. We sequenced 154 patients with plasma cell disorders and performed a head-to-head comparison with the results from conventional clinical assays, i.e., fluorescent in situ hybridization (FISH) and single-nucleotide polymorphism (SNP) microarray. Our custom capture NGS panel had high sensitivity (>99%) and specificity (>99%) for detection of IGH translocations and relevant chromosomal gains and losses in multiple myeloma. In addition, the assay was able to capture novel genomic markers associated with poor outcome such as bi-allelic events involving TP53. In summary, we show that a multiple myeloma designed custom capture NGS panel can detect IGH translocations and CNAs with very high concordance in relation to FISH and SNP microarrays and importantly captures the most relevant and recurrent somatic mutations in multiple myeloma rendering this approach highly suitable for clinical application in the modern era.
中文翻译:

全面检测复发性基因组异常:多发性骨髓瘤的靶向测序方法。
最近在多发性骨髓瘤中的基因组研究工作已经揭示了超越常规细胞遗传学分类的临床相关分子亚组。在临床试验设计和常规患者护理中实现这些进步需要新一代的分子诊断工具。在这里,我们介绍了一个定制的捕获下一代测序(NGS)面板,旨在鉴定涉及IGH基因座,臂水平和病灶拷贝数畸变的重排,以及在一次测定中多发性骨髓瘤中频繁突变的基因。我们对154例浆细胞异常患者进行了测序,并与常规临床分析(即荧光原位杂交(FISH)和单核苷酸多态性(SNP)微阵列)的结果进行了正面对比。我们的自定义捕获NGS面板具有很高的灵敏度(> 99%)和特异性(> 99%)用于检测多发性骨髓瘤中的IGH易位以及相关的染色体得失。另外,该测定法能够捕获与不良结果相关的新型基因组标记,例如涉及TP53的双等位基因事件。总而言之,我们表明,多发性骨髓瘤设计的定制捕获NGS面板可以检测与FISH和SNP芯片相关的IGH易位和CNA,并且一致性非常高,并且重要的是捕获了多发性骨髓瘤中最相关和复发的体细胞突变,因此该方法非常适合在现代临床应用。该测定法能够捕获与不良结果(例如涉及TP53的双等位基因事件)相关的新型基因组标记。总而言之,我们表明,多发性骨髓瘤设计的定制捕获NGS面板可以检测与FISH和SNP芯片相关的IGH易位和CNA,并且一致性非常高,并且重要的是捕获了多发性骨髓瘤中最相关和复发的体细胞突变,因此该方法非常适合在现代临床应用。该测定法能够捕获与不良结果(例如涉及TP53的双等位基因事件)相关的新型基因组标记。总而言之,我们表明,多发性骨髓瘤设计的定制捕获NGS面板可以检测到与FISH和SNP芯片相关的非常高一致性的IGH易位和CNA,并且重要的是捕获了多发性骨髓瘤中最相关和复发的体细胞突变,从而使该方法非常适合在现代临床应用。
更新日期:2019-12-11
中文翻译:

全面检测复发性基因组异常:多发性骨髓瘤的靶向测序方法。
最近在多发性骨髓瘤中的基因组研究工作已经揭示了超越常规细胞遗传学分类的临床相关分子亚组。在临床试验设计和常规患者护理中实现这些进步需要新一代的分子诊断工具。在这里,我们介绍了一个定制的捕获下一代测序(NGS)面板,旨在鉴定涉及IGH基因座,臂水平和病灶拷贝数畸变的重排,以及在一次测定中多发性骨髓瘤中频繁突变的基因。我们对154例浆细胞异常患者进行了测序,并与常规临床分析(即荧光原位杂交(FISH)和单核苷酸多态性(SNP)微阵列)的结果进行了正面对比。我们的自定义捕获NGS面板具有很高的灵敏度(> 99%)和特异性(> 99%)用于检测多发性骨髓瘤中的IGH易位以及相关的染色体得失。另外,该测定法能够捕获与不良结果相关的新型基因组标记,例如涉及TP53的双等位基因事件。总而言之,我们表明,多发性骨髓瘤设计的定制捕获NGS面板可以检测与FISH和SNP芯片相关的IGH易位和CNA,并且一致性非常高,并且重要的是捕获了多发性骨髓瘤中最相关和复发的体细胞突变,因此该方法非常适合在现代临床应用。该测定法能够捕获与不良结果(例如涉及TP53的双等位基因事件)相关的新型基因组标记。总而言之,我们表明,多发性骨髓瘤设计的定制捕获NGS面板可以检测与FISH和SNP芯片相关的IGH易位和CNA,并且一致性非常高,并且重要的是捕获了多发性骨髓瘤中最相关和复发的体细胞突变,因此该方法非常适合在现代临床应用。该测定法能够捕获与不良结果(例如涉及TP53的双等位基因事件)相关的新型基因组标记。总而言之,我们表明,多发性骨髓瘤设计的定制捕获NGS面板可以检测到与FISH和SNP芯片相关的非常高一致性的IGH易位和CNA,并且重要的是捕获了多发性骨髓瘤中最相关和复发的体细胞突变,从而使该方法非常适合在现代临床应用。

























 京公网安备 11010802027423号
京公网安备 11010802027423号