当前位置:
X-MOL 学术
›
PLOS Genet.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
A candidate gene analysis and GWAS for genes associated with maternal nondisjunction of chromosome 21.
PLOS Genetics ( IF 4.5 ) Pub Date : 2019-12-12 , DOI: 10.1371/journal.pgen.1008414 Jonathan M Chernus 1 , Emily G Allen 2 , Zhen Zeng 3 , Eva R Hoffman 4 , Terry J Hassold 5 , Eleanor Feingold 1, 3 , Stephanie L Sherman 2
PLOS Genetics ( IF 4.5 ) Pub Date : 2019-12-12 , DOI: 10.1371/journal.pgen.1008414 Jonathan M Chernus 1 , Emily G Allen 2 , Zhen Zeng 3 , Eva R Hoffman 4 , Terry J Hassold 5 , Eleanor Feingold 1, 3 , Stephanie L Sherman 2
Affiliation
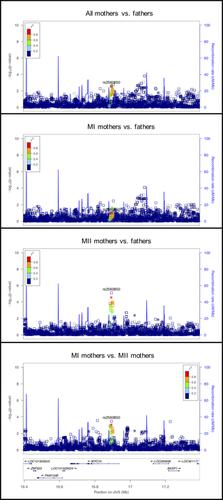
|
Human nondisjunction errors in oocytes are the leading cause of pregnancy loss, and for pregnancies that continue to term, the leading cause of intellectual disabilities and birth defects. For the first time, we have conducted a candidate gene and genome-wide association study to identify genes associated with maternal nondisjunction of chromosome 21 as a first step to understand predisposing factors. A total of 2,186 study participants were genotyped on the HumanOmniExpressExome-8v1-2 array. These participants included 749 live birth offspring with standard trisomy 21 and 1,437 parents. Genotypes from the parents and child were then used to identify mothers with nondisjunction errors derived in the oocyte and to establish the type of error (meiosis I or meiosis II). We performed a unique set of subgroup comparisons designed to leverage our previous work suggesting that the etiologies of meiosis I and meiosis II nondisjunction differ for trisomy 21. For the candidate gene analysis, we selected genes associated with chromosome dynamics early in meiosis and genes associated with human global recombination counts. Several candidate genes showed strong associations with maternal nondisjunction of chromosome 21, demonstrating that genetic variants associated with normal variation in meiotic processes can be risk factors for nondisjunction. The genome-wide analysis also suggested several new potentially associated loci, although follow-up studies using independent samples are required.
中文翻译:

候选基因分析和GWAS,用于与21号染色体的孕妇非分离相关的基因。
卵母细胞中的人类非分离错误是导致妊娠流产的主要原因,而对于持续妊娠的妊娠,则是智力障碍和出生缺陷的主要原因。首次,我们进行了候选基因和全基因组关联研究,以鉴定与21号染色体母系非分离相关的基因,这是了解易患因素的第一步。在HumanOmniExpressExome-8v1-2阵列上对2186位研究参与者进行了基因分型。这些参与者包括749个标准21三体动物的活产后代和1,437个父母。然后使用来自父母和孩子的基因型来鉴定在卵母细胞中具有非分离错误的母亲,并确定错误的类型(减数分裂I或减数分裂II)。我们进行了一组独特的亚组比较,目的是利用我们以前的工作,这表明21三体性的减数分裂I和减数分裂II非分离的病因学有所不同。对于候选基因分析,我们选择了与减数分裂早期的染色体动力学相关的基因以及与减数分裂相关的基因。人类全球重组计数。几个候选基因显示出与21号染色体的母体非分离性密切相关,表明与减数分裂过程中正常变异相关的遗传变异可能是非分离性的危险因素。尽管需要使用独立样本进行后续研究,但全基因组分析还提出了几个新的潜在相关基因座。对于候选基因分析,我们选择了与减数分裂早期染色体动态相关的基因和与人类整体重组计数相关的基因。几个候选基因显示出与21号染色体的母体非分离性密切相关,表明与减数分裂过程中正常变异相关的遗传变异可能是非分离性的危险因素。尽管需要使用独立样本进行后续研究,但全基因组分析还提出了几个新的潜在相关基因座。对于候选基因分析,我们选择了与减数分裂早期染色体动态相关的基因和与人类整体重组计数相关的基因。几个候选基因显示出与21号染色体的母体非分离性密切相关,表明与减数分裂过程中正常变异相关的遗传变异可能是非分离性的危险因素。尽管需要使用独立样本进行后续研究,但全基因组分析还提出了几个新的潜在相关基因座。证明与减数分裂过程正常变异相关的遗传变异可能是不分离的危险因素。尽管需要使用独立样本进行后续研究,但全基因组分析还提出了几个新的潜在相关基因座。证明与减数分裂过程正常变异相关的遗传变异可能是不分离的危险因素。尽管需要使用独立样本进行后续研究,但全基因组分析还提出了几个新的潜在相关基因座。
更新日期:2019-12-13
中文翻译:

候选基因分析和GWAS,用于与21号染色体的孕妇非分离相关的基因。
卵母细胞中的人类非分离错误是导致妊娠流产的主要原因,而对于持续妊娠的妊娠,则是智力障碍和出生缺陷的主要原因。首次,我们进行了候选基因和全基因组关联研究,以鉴定与21号染色体母系非分离相关的基因,这是了解易患因素的第一步。在HumanOmniExpressExome-8v1-2阵列上对2186位研究参与者进行了基因分型。这些参与者包括749个标准21三体动物的活产后代和1,437个父母。然后使用来自父母和孩子的基因型来鉴定在卵母细胞中具有非分离错误的母亲,并确定错误的类型(减数分裂I或减数分裂II)。我们进行了一组独特的亚组比较,目的是利用我们以前的工作,这表明21三体性的减数分裂I和减数分裂II非分离的病因学有所不同。对于候选基因分析,我们选择了与减数分裂早期的染色体动力学相关的基因以及与减数分裂相关的基因。人类全球重组计数。几个候选基因显示出与21号染色体的母体非分离性密切相关,表明与减数分裂过程中正常变异相关的遗传变异可能是非分离性的危险因素。尽管需要使用独立样本进行后续研究,但全基因组分析还提出了几个新的潜在相关基因座。对于候选基因分析,我们选择了与减数分裂早期染色体动态相关的基因和与人类整体重组计数相关的基因。几个候选基因显示出与21号染色体的母体非分离性密切相关,表明与减数分裂过程中正常变异相关的遗传变异可能是非分离性的危险因素。尽管需要使用独立样本进行后续研究,但全基因组分析还提出了几个新的潜在相关基因座。对于候选基因分析,我们选择了与减数分裂早期染色体动态相关的基因和与人类整体重组计数相关的基因。几个候选基因显示出与21号染色体的母体非分离性密切相关,表明与减数分裂过程中正常变异相关的遗传变异可能是非分离性的危险因素。尽管需要使用独立样本进行后续研究,但全基因组分析还提出了几个新的潜在相关基因座。证明与减数分裂过程正常变异相关的遗传变异可能是不分离的危险因素。尽管需要使用独立样本进行后续研究,但全基因组分析还提出了几个新的潜在相关基因座。证明与减数分裂过程正常变异相关的遗传变异可能是不分离的危险因素。尽管需要使用独立样本进行后续研究,但全基因组分析还提出了几个新的潜在相关基因座。



























 京公网安备 11010802027423号
京公网安备 11010802027423号