当前位置:
X-MOL 学术
›
Br. J. Cancer
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Kinetic modelling of quantitative proteome data predicts metabolic reprogramming of liver cancer.
British Journal of Cancer ( IF 8.8 ) Pub Date : 2019-12-10 , DOI: 10.1038/s41416-019-0659-3 Nikolaus Berndt 1, 2 , Antje Egners 3 , Guido Mastrobuoni 4 , Olga Vvedenskaya 4 , Athanassios Fragoulis 3 , Aurélien Dugourd 5 , Sascha Bulik 1 , Matthias Pietzke 4 , Chris Bielow 4, 6 , Rob van Gassel 7, 8 , Steven W Olde Damink 7, 8, 9, 10, 11 , Merve Erdem 3 , Julio Saez-Rodriguez 5 , Hermann-Georg Holzhütter 1 , Stefan Kempa 4 , Thorsten Cramer 3, 7, 8, 10, 11
British Journal of Cancer ( IF 8.8 ) Pub Date : 2019-12-10 , DOI: 10.1038/s41416-019-0659-3 Nikolaus Berndt 1, 2 , Antje Egners 3 , Guido Mastrobuoni 4 , Olga Vvedenskaya 4 , Athanassios Fragoulis 3 , Aurélien Dugourd 5 , Sascha Bulik 1 , Matthias Pietzke 4 , Chris Bielow 4, 6 , Rob van Gassel 7, 8 , Steven W Olde Damink 7, 8, 9, 10, 11 , Merve Erdem 3 , Julio Saez-Rodriguez 5 , Hermann-Georg Holzhütter 1 , Stefan Kempa 4 , Thorsten Cramer 3, 7, 8, 10, 11
Affiliation
|
BACKGROUND
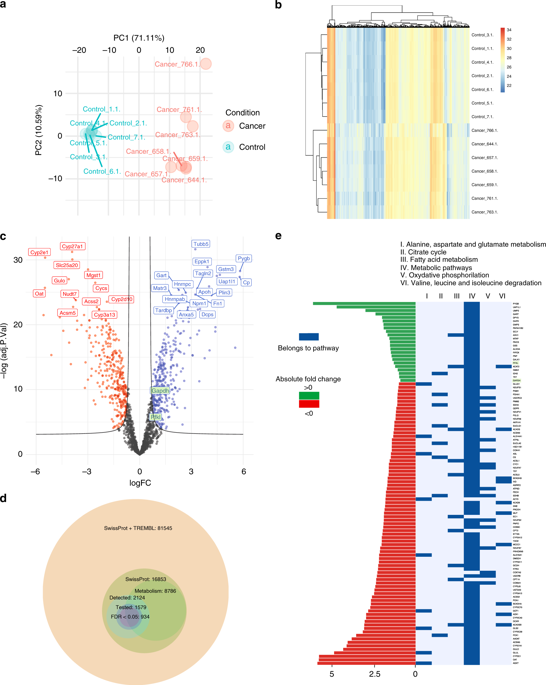
Metabolic alterations can serve as targets for diagnosis and cancer therapy. Due to the highly complex regulation of cellular metabolism, definite identification of metabolic pathway alterations remains challenging and requires sophisticated experimentation.
METHODS
We applied a comprehensive kinetic model of the central carbon metabolism (CCM) to characterise metabolic reprogramming in murine liver cancer.
RESULTS
We show that relative differences of protein abundances of metabolic enzymes obtained by mass spectrometry can be used to assess their maximal velocity values. Model simulations predicted tumour-specific alterations of various components of the CCM, a selected number of which were subsequently verified by in vitro and in vivo experiments. Furthermore, we demonstrate the ability of the kinetic model to identify metabolic pathways whose inhibition results in selective tumour cell killing.
CONCLUSIONS
Our systems biology approach establishes that combining cellular experimentation with computer simulations of physiology-based metabolic models enables a comprehensive understanding of deregulated energetics in cancer. We propose that modelling proteomics data from human HCC with our approach will enable an individualised metabolic profiling of tumours and predictions of the efficacy of drug therapies targeting specific metabolic pathways.
中文翻译:

定量蛋白质组数据的动力学模型预测肝癌的代谢重编程。
背景代谢改变可以作为诊断和癌症治疗的目标。由于细胞代谢的高度复杂的调节,代谢途径改变的明确识别仍然具有挑战性,需要复杂的实验。方法 我们应用中央碳代谢 (CCM) 的综合动力学模型来表征小鼠肝癌的代谢重编程。结果我们表明,通过质谱法获得的代谢酶蛋白质丰度的相对差异可用于评估它们的最大速度值。模型模拟预测了 CCM 各种成分的肿瘤特异性改变,随后通过体外和体内实验验证了其中一些成分。此外,我们证明了动力学模型能够识别抑制导致选择性肿瘤细胞杀伤的代谢途径。结论 我们的系统生物学方法表明,将细胞实验与基于生理学的代谢模型的计算机模拟相结合,可以全面了解癌症中失调的能量学。我们建议使用我们的方法对来自人类 HCC 的蛋白质组学数据进行建模,这将能够对肿瘤进行个体化的代谢分析,并预测针对特定代谢途径的药物疗法的功效。结论 我们的系统生物学方法表明,将细胞实验与基于生理学的代谢模型的计算机模拟相结合,可以全面了解癌症中失调的能量学。我们建议使用我们的方法对来自人类 HCC 的蛋白质组学数据进行建模,这将能够对肿瘤进行个体化的代谢分析,并预测针对特定代谢途径的药物疗法的功效。结论 我们的系统生物学方法表明,将细胞实验与基于生理学的代谢模型的计算机模拟相结合,可以全面了解癌症中失调的能量学。我们建议使用我们的方法对来自人类 HCC 的蛋白质组学数据进行建模,这将能够对肿瘤进行个体化的代谢分析,并预测针对特定代谢途径的药物疗法的功效。
更新日期:2019-12-11
中文翻译:

定量蛋白质组数据的动力学模型预测肝癌的代谢重编程。
背景代谢改变可以作为诊断和癌症治疗的目标。由于细胞代谢的高度复杂的调节,代谢途径改变的明确识别仍然具有挑战性,需要复杂的实验。方法 我们应用中央碳代谢 (CCM) 的综合动力学模型来表征小鼠肝癌的代谢重编程。结果我们表明,通过质谱法获得的代谢酶蛋白质丰度的相对差异可用于评估它们的最大速度值。模型模拟预测了 CCM 各种成分的肿瘤特异性改变,随后通过体外和体内实验验证了其中一些成分。此外,我们证明了动力学模型能够识别抑制导致选择性肿瘤细胞杀伤的代谢途径。结论 我们的系统生物学方法表明,将细胞实验与基于生理学的代谢模型的计算机模拟相结合,可以全面了解癌症中失调的能量学。我们建议使用我们的方法对来自人类 HCC 的蛋白质组学数据进行建模,这将能够对肿瘤进行个体化的代谢分析,并预测针对特定代谢途径的药物疗法的功效。结论 我们的系统生物学方法表明,将细胞实验与基于生理学的代谢模型的计算机模拟相结合,可以全面了解癌症中失调的能量学。我们建议使用我们的方法对来自人类 HCC 的蛋白质组学数据进行建模,这将能够对肿瘤进行个体化的代谢分析,并预测针对特定代谢途径的药物疗法的功效。结论 我们的系统生物学方法表明,将细胞实验与基于生理学的代谢模型的计算机模拟相结合,可以全面了解癌症中失调的能量学。我们建议使用我们的方法对来自人类 HCC 的蛋白质组学数据进行建模,这将能够对肿瘤进行个体化的代谢分析,并预测针对特定代谢途径的药物疗法的功效。



























 京公网安备 11010802027423号
京公网安备 11010802027423号