当前位置:
X-MOL 学术
›
Nat. Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Accurate annotation of human protein-coding small open reading frames.
Nature Chemical Biology ( IF 14.8 ) Pub Date : 2019-12-09 , DOI: 10.1038/s41589-019-0425-0 Thomas F Martinez 1 , Qian Chu 1 , Cynthia Donaldson 1 , Dan Tan 1 , Maxim N Shokhirev 2 , Alan Saghatelian 1
Nature Chemical Biology ( IF 14.8 ) Pub Date : 2019-12-09 , DOI: 10.1038/s41589-019-0425-0 Thomas F Martinez 1 , Qian Chu 1 , Cynthia Donaldson 1 , Dan Tan 1 , Maxim N Shokhirev 2 , Alan Saghatelian 1
Affiliation
|
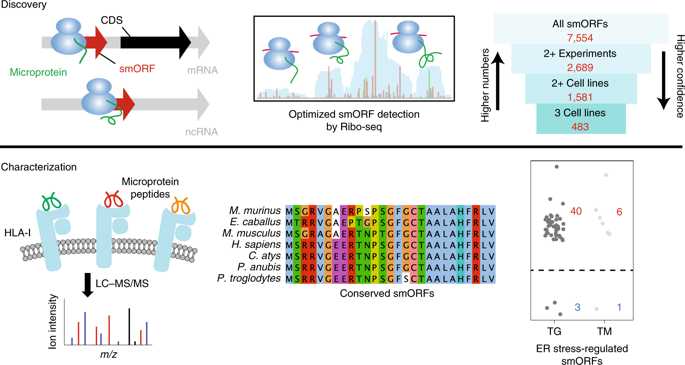
Functional protein-coding small open reading frames (smORFs) are emerging as an important class of genes. However, the number of translated smORFs in the human genome is unclear because proteogenomic methods are not sensitive enough, and, as we show, Ribo-seq strategies require additional measures to ensure comprehensive and accurate smORF annotation. Here, we integrate de novo transcriptome assembly and Ribo-seq into an improved workflow that overcomes obstacles with previous methods, to more confidently annotate thousands of smORFs. Evolutionary conservation analyses suggest that hundreds of smORF-encoded microproteins are likely functional. Additionally, many smORFs are regulated during fundamental biological processes, such as cell stress. Peptides derived from smORFs are also detectable on human leukocyte antigen complexes, revealing smORFs as a source of antigens. Thus, by including additional validation into our smORF annotation workflow, we accurately identify thousands of unannotated translated smORFs that will provide a rich pool of unexplored, functional human genes.
中文翻译:

人类蛋白质编码小开放阅读框的准确注释。
功能性蛋白质编码小开放阅读框(smORF)正在成为一类重要的基因。然而,人类基因组中翻译的 smORF 数量尚不清楚,因为蛋白质组学方法不够灵敏,而且正如我们所表明的,Ribo-seq 策略需要额外的措施来确保全面且准确的 smORF 注释。在这里,我们将从头转录组组装和 Ribo-seq 集成到改进的工作流程中,克服了以前方法的障碍,更自信地注释数千个 smORF。进化保守分析表明,数百种 smORF 编码的微生物蛋白可能具有功能。此外,许多 smORF 在细胞应激等基本生物过程中受到调节。源自 smORF 的肽也可在人类白细胞抗原复合物上检测到,揭示了 smORF 作为抗原的来源。因此,通过在我们的 smORF 注释工作流程中加入额外的验证,我们可以准确识别数千个未注释的翻译 smORF,这将提供丰富的未探索的功能性人类基因库。
更新日期:2019-12-11
中文翻译:

人类蛋白质编码小开放阅读框的准确注释。
功能性蛋白质编码小开放阅读框(smORF)正在成为一类重要的基因。然而,人类基因组中翻译的 smORF 数量尚不清楚,因为蛋白质组学方法不够灵敏,而且正如我们所表明的,Ribo-seq 策略需要额外的措施来确保全面且准确的 smORF 注释。在这里,我们将从头转录组组装和 Ribo-seq 集成到改进的工作流程中,克服了以前方法的障碍,更自信地注释数千个 smORF。进化保守分析表明,数百种 smORF 编码的微生物蛋白可能具有功能。此外,许多 smORF 在细胞应激等基本生物过程中受到调节。源自 smORF 的肽也可在人类白细胞抗原复合物上检测到,揭示了 smORF 作为抗原的来源。因此,通过在我们的 smORF 注释工作流程中加入额外的验证,我们可以准确识别数千个未注释的翻译 smORF,这将提供丰富的未探索的功能性人类基因库。


























 京公网安备 11010802027423号
京公网安备 11010802027423号