当前位置:
X-MOL 学术
›
Mol. Psychiatry
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Epigenome-wide meta-analysis of blood DNA methylation and its association with subcortical volumes: findings from the ENIGMA Epigenetics Working Group.
Molecular Psychiatry ( IF 11.0 ) Pub Date : 2019-12-06 , DOI: 10.1038/s41380-019-0605-z Tianye Jia 1, 2, 3 , Congying Chu 1 , Yun Liu 4 , Jenny van Dongen 5 , Evangelos Papastergios 1 , Nicola J Armstrong 6 , Mark E Bastin 7 , Tania Carrillo-Roa 8 , Anouk den Braber 5 , Mathew Harris 9 , Rick Jansen 10 , Jingyu Liu 11 , Michelle Luciano 12 , Anil P S Ori 13 , Roberto Roiz Santiañez 14, 15 , Barbara Ruggeri 1 , Daniil Sarkisyan 16 , Jean Shin 17 , Kim Sungeun 18, 19 , Diana Tordesillas Gutiérrez 14, 20 , Dennis Van't Ent 5 , David Ames 21, 22 , Eric Artiges 23, 24, 25 , Georgy Bakalkin 16 , Tobias Banaschewski 26 , Arun L W Bokde 27 , Henry Brodaty 28, 29 , Uli Bromberg 30 , Rachel Brouwer 31 , Christian Büchel 30 , Erin Burke Quinlan 1 , Wiepke Cahn 31 , Greig I de Zubicaray 32 , Stefan Ehrlich 33 , Tomas J Ekström 34 , Herta Flor 35, 36 , Juliane H Fröhner 37 , Vincent Frouin 38 , Hugh Garavan 39 , Penny Gowland 40 , Andreas Heinz 41, 42 , Jacqueline Hoare 43 , Bernd Ittermann 44 , Neda Jahanshad 45 , Jiyang Jiang 28 , John B Kwok 46, 47 , Nicholas G Martin 48 , Jean-Luc Martinot 23, 24, 49 , Karen A Mather 28, 50 , Katie L McMahon 51 , Allan F McRae 52 , Frauke Nees 26, 35 , Dimitri Papadopoulos Orfanos 38 , Tomáš Paus 53 , Luise Poustka 54 , Philipp G Sämann 8 , Peter R Schofield 50, 55 , Michael N Smolka 37 , Dan J Stein 43, 56 , Lachlan T Strike 57 , Jalmar Teeuw 13, 31 , Anbupalam Thalamuthu 28, 50 , Julian Trollor 28, 58 , Henrik Walter 41, 42 , Joanna M Wardlaw 59, 60 , Wei Wen 28 , Robert Whelan 61 , Liana G Apostolova 62, 63, 64, 65 , Elisabeth B Binder 8 , Dorret I Boomsma 5 , Vince Calhoun 11, 66 , Benedicto Crespo-Facorro 14 , Ian J Deary 12 , Hilleke Hulshoff Pol 31 , Roel A Ophoff 31, 67 , Zdenka Pausova 17 , Perminder S Sachdev 28, 68 , Andrew Saykin 69 , Margaret J Wright 48 , Paul M Thompson 45 , Gunter Schumann 1 , Sylvane Desrivières 1
Molecular Psychiatry ( IF 11.0 ) Pub Date : 2019-12-06 , DOI: 10.1038/s41380-019-0605-z Tianye Jia 1, 2, 3 , Congying Chu 1 , Yun Liu 4 , Jenny van Dongen 5 , Evangelos Papastergios 1 , Nicola J Armstrong 6 , Mark E Bastin 7 , Tania Carrillo-Roa 8 , Anouk den Braber 5 , Mathew Harris 9 , Rick Jansen 10 , Jingyu Liu 11 , Michelle Luciano 12 , Anil P S Ori 13 , Roberto Roiz Santiañez 14, 15 , Barbara Ruggeri 1 , Daniil Sarkisyan 16 , Jean Shin 17 , Kim Sungeun 18, 19 , Diana Tordesillas Gutiérrez 14, 20 , Dennis Van't Ent 5 , David Ames 21, 22 , Eric Artiges 23, 24, 25 , Georgy Bakalkin 16 , Tobias Banaschewski 26 , Arun L W Bokde 27 , Henry Brodaty 28, 29 , Uli Bromberg 30 , Rachel Brouwer 31 , Christian Büchel 30 , Erin Burke Quinlan 1 , Wiepke Cahn 31 , Greig I de Zubicaray 32 , Stefan Ehrlich 33 , Tomas J Ekström 34 , Herta Flor 35, 36 , Juliane H Fröhner 37 , Vincent Frouin 38 , Hugh Garavan 39 , Penny Gowland 40 , Andreas Heinz 41, 42 , Jacqueline Hoare 43 , Bernd Ittermann 44 , Neda Jahanshad 45 , Jiyang Jiang 28 , John B Kwok 46, 47 , Nicholas G Martin 48 , Jean-Luc Martinot 23, 24, 49 , Karen A Mather 28, 50 , Katie L McMahon 51 , Allan F McRae 52 , Frauke Nees 26, 35 , Dimitri Papadopoulos Orfanos 38 , Tomáš Paus 53 , Luise Poustka 54 , Philipp G Sämann 8 , Peter R Schofield 50, 55 , Michael N Smolka 37 , Dan J Stein 43, 56 , Lachlan T Strike 57 , Jalmar Teeuw 13, 31 , Anbupalam Thalamuthu 28, 50 , Julian Trollor 28, 58 , Henrik Walter 41, 42 , Joanna M Wardlaw 59, 60 , Wei Wen 28 , Robert Whelan 61 , Liana G Apostolova 62, 63, 64, 65 , Elisabeth B Binder 8 , Dorret I Boomsma 5 , Vince Calhoun 11, 66 , Benedicto Crespo-Facorro 14 , Ian J Deary 12 , Hilleke Hulshoff Pol 31 , Roel A Ophoff 31, 67 , Zdenka Pausova 17 , Perminder S Sachdev 28, 68 , Andrew Saykin 69 , Margaret J Wright 48 , Paul M Thompson 45 , Gunter Schumann 1 , Sylvane Desrivières 1
Affiliation
|
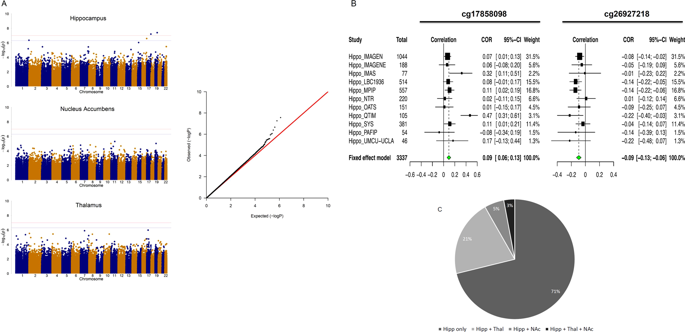
DNA methylation, which is modulated by both genetic factors and environmental exposures, may offer a unique opportunity to discover novel biomarkers of disease-related brain phenotypes, even when measured in other tissues than brain, such as blood. A few studies of small sample sizes have revealed associations between blood DNA methylation and neuropsychopathology, however, large-scale epigenome-wide association studies (EWAS) are needed to investigate the utility of DNA methylation profiling as a peripheral marker for the brain. Here, in an analysis of eleven international cohorts, totalling 3337 individuals, we report epigenome-wide meta-analyses of blood DNA methylation with volumes of the hippocampus, thalamus and nucleus accumbens (NAcc)-three subcortical regions selected for their associations with disease and heritability and volumetric variability. Analyses of individual CpGs revealed genome-wide significant associations with hippocampal volume at two loci. No significant associations were found for analyses of thalamus and nucleus accumbens volumes. Cluster-based analyses revealed additional differentially methylated regions (DMRs) associated with hippocampal volume. DNA methylation at these loci affected expression of proximal genes involved in learning and memory, stem cell maintenance and differentiation, fatty acid metabolism and type-2 diabetes. These DNA methylation marks, their interaction with genetic variants and their impact on gene expression offer new insights into the relationship between epigenetic variation and brain structure and may provide the basis for biomarker discovery in neurodegeneration and neuropsychiatric conditions.
中文翻译:

血液 DNA 甲基化的全表观基因组荟萃分析及其与皮层下体积的关联:来自 ENIGMA 表观遗传学工作组的发现。
受遗传因素和环境暴露调节的 DNA 甲基化可能提供独特的机会来发现与疾病相关的大脑表型的新生物标志物,即使在大脑以外的其他组织(如血液)中进行测量也是如此。一些小样本研究揭示了血液 DNA 甲基化与神经精神病理学之间的关联,然而,需要大规模表观基因组范围的关联研究 (EWAS) 来研究 DNA 甲基化分析作为大脑外周标记的效用。在这里,在对 11 个国际队列(共 3337 人)的分析中,我们报告了血液 DNA 甲基化与海马体积的表观基因组范围的荟萃分析,丘脑和伏隔核 (NAcc) - 三个皮层下区域因其与疾病和遗传力以及体积变异性的关联而被选中。对单个 CpG 的分析揭示了全基因组与两个位点的海马体积显着相关。对于丘脑和伏隔核体积的分析没有发现显着关联。基于聚类的分析揭示了与海马体积相关的其他差异甲基化区域 (DMR)。这些位点的 DNA 甲基化影响了参与学习和记忆、干细胞维持和分化、脂肪酸代谢和 2 型糖尿病的近端基因的表达。这些DNA甲基化标记,
更新日期:2019-12-06
中文翻译:

血液 DNA 甲基化的全表观基因组荟萃分析及其与皮层下体积的关联:来自 ENIGMA 表观遗传学工作组的发现。
受遗传因素和环境暴露调节的 DNA 甲基化可能提供独特的机会来发现与疾病相关的大脑表型的新生物标志物,即使在大脑以外的其他组织(如血液)中进行测量也是如此。一些小样本研究揭示了血液 DNA 甲基化与神经精神病理学之间的关联,然而,需要大规模表观基因组范围的关联研究 (EWAS) 来研究 DNA 甲基化分析作为大脑外周标记的效用。在这里,在对 11 个国际队列(共 3337 人)的分析中,我们报告了血液 DNA 甲基化与海马体积的表观基因组范围的荟萃分析,丘脑和伏隔核 (NAcc) - 三个皮层下区域因其与疾病和遗传力以及体积变异性的关联而被选中。对单个 CpG 的分析揭示了全基因组与两个位点的海马体积显着相关。对于丘脑和伏隔核体积的分析没有发现显着关联。基于聚类的分析揭示了与海马体积相关的其他差异甲基化区域 (DMR)。这些位点的 DNA 甲基化影响了参与学习和记忆、干细胞维持和分化、脂肪酸代谢和 2 型糖尿病的近端基因的表达。这些DNA甲基化标记,



























 京公网安备 11010802027423号
京公网安备 11010802027423号