当前位置:
X-MOL 学术
›
J. Comput. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
PepPro: A Nonredundant Structure Data Set for Benchmarking Peptide–Protein Computational Docking
Journal of Computational Chemistry ( IF 3 ) Pub Date : 2019-12-02 , DOI: 10.1002/jcc.26114 Xianjin Xu 1, 2, 3, 4 , Xiaoqin Zou 1, 2, 3, 4
Journal of Computational Chemistry ( IF 3 ) Pub Date : 2019-12-02 , DOI: 10.1002/jcc.26114 Xianjin Xu 1, 2, 3, 4 , Xiaoqin Zou 1, 2, 3, 4
Affiliation
|
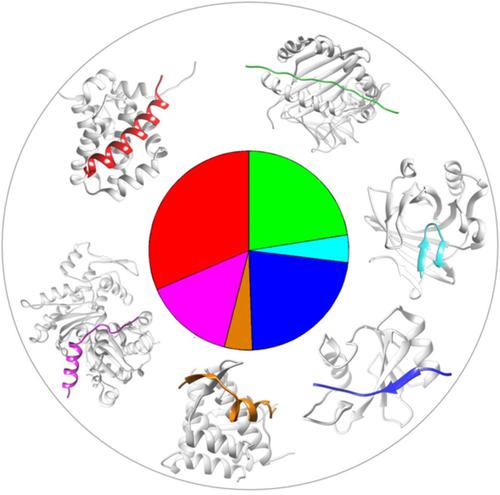
We present a nonredundant benchmark, coined PepPro, for testing peptide–protein docking algorithms. Currently, PepPro contains 89 nonredundant experimentally determined peptide–protein complex structures, with peptide sequence lengths ranging from 5 to 30 amino acids. The benchmark covers peptides with distinct secondary structures, including helix, partial helix, a mixture of helix and β‐sheet, β‐sheet formed through binding, β‐sheet formed through self‐folding, and coil. In addition, unbound proteins' structures are provided for 58 complexes and can be used for testing the ability of a docking algorithm handling the conformational changes of proteins during the binding process. PepPro should benefit the docking community for the development and improvement of peptide docking algorithms. The benchmark is available at http://zoulab.dalton.missouri.edu/PepPro_benchmark. © 2019 Wiley Periodicals, Inc.
中文翻译:

PepPro:用于基准肽-蛋白质计算对接的非冗余结构数据集
我们提出了一个非冗余基准,创造了 PepPro,用于测试肽-蛋白质对接算法。目前,PepPro 包含 89 个非冗余的实验确定的肽-蛋白质复合结构,肽序列长度为 5 到 30 个氨基酸。该基准涵盖具有不同二级结构的肽,包括螺旋、部分螺旋、螺旋和β-折叠的混合物、通过结合形成的β-折叠、通过自折叠形成的β-折叠和卷曲。此外,还提供了 58 个复合物的未结合蛋白质结构,可用于测试对接算法在结合过程中处理蛋白质构象变化的能力。PepPro 应该使对接社区受益,以开发和改进肽对接算法。该基准可从 http://zoulab.dalton 获得。Missouri.edu/PepPro_benchmark。© 2019 威利期刊公司。
更新日期:2019-12-02
中文翻译:

PepPro:用于基准肽-蛋白质计算对接的非冗余结构数据集
我们提出了一个非冗余基准,创造了 PepPro,用于测试肽-蛋白质对接算法。目前,PepPro 包含 89 个非冗余的实验确定的肽-蛋白质复合结构,肽序列长度为 5 到 30 个氨基酸。该基准涵盖具有不同二级结构的肽,包括螺旋、部分螺旋、螺旋和β-折叠的混合物、通过结合形成的β-折叠、通过自折叠形成的β-折叠和卷曲。此外,还提供了 58 个复合物的未结合蛋白质结构,可用于测试对接算法在结合过程中处理蛋白质构象变化的能力。PepPro 应该使对接社区受益,以开发和改进肽对接算法。该基准可从 http://zoulab.dalton 获得。Missouri.edu/PepPro_benchmark。© 2019 威利期刊公司。


























 京公网安备 11010802027423号
京公网安备 11010802027423号