当前位置:
X-MOL 学术
›
Nat. Genet.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Histone hyperacetylation disrupts core gene regulatory architecture in rhabdomyosarcoma.
Nature Genetics ( IF 30.8 ) Pub Date : 2019-11-29 , DOI: 10.1038/s41588-019-0534-4 Berkley E Gryder 1 , Silvia Pomella 1, 2 , Carly Sayers 3 , Xiaoli S Wu 4, 5 , Young Song 1 , Anna M Chiarella 6 , Sukriti Bagchi 3 , Hsien-Chao Chou 1 , Ranu S Sinniah 1 , Ashley Walton 1 , Xinyu Wen 1, 7 , Rossella Rota 2 , Nathaniel A Hathaway 6 , Keji Zhao 8 , Jiji Chen 9 , Christopher R Vakoc 4 , Jack F Shern 3 , Benjamin Z Stanton 1, 10 , Javed Khan 1
Nature Genetics ( IF 30.8 ) Pub Date : 2019-11-29 , DOI: 10.1038/s41588-019-0534-4 Berkley E Gryder 1 , Silvia Pomella 1, 2 , Carly Sayers 3 , Xiaoli S Wu 4, 5 , Young Song 1 , Anna M Chiarella 6 , Sukriti Bagchi 3 , Hsien-Chao Chou 1 , Ranu S Sinniah 1 , Ashley Walton 1 , Xinyu Wen 1, 7 , Rossella Rota 2 , Nathaniel A Hathaway 6 , Keji Zhao 8 , Jiji Chen 9 , Christopher R Vakoc 4 , Jack F Shern 3 , Benjamin Z Stanton 1, 10 , Javed Khan 1
Affiliation
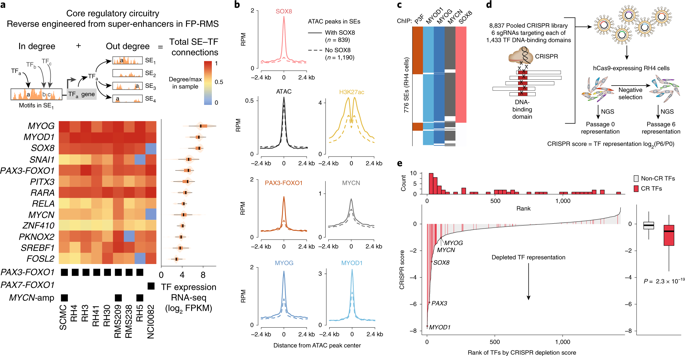
|
Core regulatory transcription factors (CR TFs) orchestrate the placement of super-enhancers (SEs) to activate transcription of cell-identity specifying gene networks, and are critical in promoting cancer. Here, we define the core regulatory circuitry of rhabdomyosarcoma and identify critical CR TF dependencies. These CR TFs build SEs that have the highest levels of histone acetylation, yet paradoxically the same SEs also harbor the greatest amounts of histone deacetylases. We find that hyperacetylation selectively halts CR TF transcription. To investigate the architectural determinants of this phenotype, we used absolute quantification of architecture (AQuA) HiChIP, which revealed erosion of native SE contacts, and aberrant spreading of contacts that involved histone acetylation. Hyperacetylation removes RNA polymerase II (RNA Pol II) from core regulatory genetic elements, and eliminates RNA Pol II but not BRD4 phase condensates. This study identifies an SE-specific requirement for balancing histone modification states to maintain SE architecture and CR TF transcription.
中文翻译:

组蛋白高度乙酰化破坏了横纹肌肉瘤的核心基因调控结构。
核心调节转录因子 (CR TF) 协调超级增强子 (SE) 的放置以激活细胞身份指定基因网络的转录,并且在促进癌症方面至关重要。在这里,我们定义了横纹肌肉瘤的核心调节回路并确定了关键的 CR TF 依赖性。这些 CR TF 构建的 SE 具有最高水平的组蛋白乙酰化,但矛盾的是,相同的 SE 也含有最大量的组蛋白脱乙酰酶。我们发现超乙酰化选择性地停止 CR TF 转录。为了研究这种表型的结构决定因素,我们使用了结构绝对量化 (AQuA) HiChIP,它揭示了原生 SE 接触的侵蚀,以及涉及组蛋白乙酰化的接触的异常扩散。超乙酰化从核心调控遗传元件中去除 RNA 聚合酶 II (RNA Pol II),并去除 RNA Pol II 但不去除 BRD4 相凝聚物。本研究确定了平衡组蛋白修饰状态以维持 SE 结构和 CR TF 转录的 SE 特定要求。
更新日期:2019-11-30
中文翻译:

组蛋白高度乙酰化破坏了横纹肌肉瘤的核心基因调控结构。
核心调节转录因子 (CR TF) 协调超级增强子 (SE) 的放置以激活细胞身份指定基因网络的转录,并且在促进癌症方面至关重要。在这里,我们定义了横纹肌肉瘤的核心调节回路并确定了关键的 CR TF 依赖性。这些 CR TF 构建的 SE 具有最高水平的组蛋白乙酰化,但矛盾的是,相同的 SE 也含有最大量的组蛋白脱乙酰酶。我们发现超乙酰化选择性地停止 CR TF 转录。为了研究这种表型的结构决定因素,我们使用了结构绝对量化 (AQuA) HiChIP,它揭示了原生 SE 接触的侵蚀,以及涉及组蛋白乙酰化的接触的异常扩散。超乙酰化从核心调控遗传元件中去除 RNA 聚合酶 II (RNA Pol II),并去除 RNA Pol II 但不去除 BRD4 相凝聚物。本研究确定了平衡组蛋白修饰状态以维持 SE 结构和 CR TF 转录的 SE 特定要求。


























 京公网安备 11010802027423号
京公网安备 11010802027423号