当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Double-Check Base Editing for Efficient A to G Conversions.
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2019-12-02 , DOI: 10.1021/acssynbio.9b00284 Xiuqing Xin 1, 2 , Ju Li 3 , Dongdong Zhao 2 , Siwei Li 2 , Qianwen Xie 1, 2 , Zhongkang Li 2 , Feiyu Fan 2 , Changhao Bi 2 , Xueli Zhang 2
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2019-12-02 , DOI: 10.1021/acssynbio.9b00284 Xiuqing Xin 1, 2 , Ju Li 3 , Dongdong Zhao 2 , Siwei Li 2 , Qianwen Xie 1, 2 , Zhongkang Li 2 , Feiyu Fan 2 , Changhao Bi 2 , Xueli Zhang 2
Affiliation
|
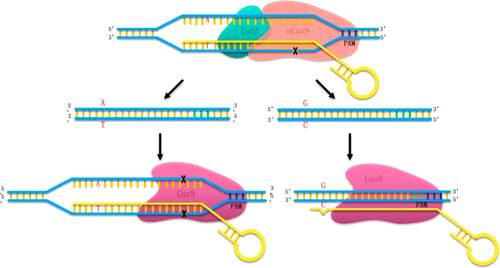
With the development of CRISPR/Cas9 technology, a new generation of editing methods that convert specific bases has enabled precise single-base mutations. To date, conversion of cytosine to thymidine and adenine to guanine has been achieved using the cytidine deaminase APOBEC1 and adenosine deaminase (TadA), respectively. However, the base editing efficiency can be unacceptably low in some cell types or at certain target loci. One reason might be the lack of a selective pressure against the survival of nonedited cells. Few studies on ABE in prokaryotes have been reported, probably due to the relatively low editing efficiency of TadA. Improving the editing efficiency is the key for establishing base editing techniques and especially the ABE technologies. In this work, a selective pressure against nonedited cells was implemented to increase the base editing efficiency. First, we fused nCas9 or dCas9 with TadA to compare the editing efficiency of nCas9-TadA and dCas9-TadA fusion complexes in the model prokaryote Escherichia coli. While nCas9-TadA was able to achieve A to G base editing (ABE) with a moderate efficiency, dCas9-TadA had a very low efficiency. To enrich for edited cells and increase the base-editing efficiency, we utilized the induction of double-strand breaks by active Cas9, which leads to the death of prokaryotic cells. By introducing an inducible active Cas9 with the same editing gRNA as the nCas9-TadA in the base editing process, the cells with nonedited target bases remained vulnerable to Cas9 and were eliminated. Thus, a double-check base editing (DBE) method was established, which significantly improved the editing efficiency of ABE in E. coli, reaching 99.0% for some sites. By placing a selective pressure against nonedited cells, the DBE strategy might also be applied to various scenarios to increase the efficiency of many different base editing targets or even for epigenetic DNA modification techniques.
中文翻译:

仔细检查基本编辑,以实现有效的A到G转换。
随着CRISPR / Cas9技术的发展,转换特定碱基的新一代编辑方法实现了精确的单碱基突变。迄今为止,已经分别使用胞苷脱氨酶APOBEC1和腺苷脱氨酶(TadA)实现了胞嘧啶向胸苷的转化和腺嘌呤向鸟嘌呤的转化。但是,在某些细胞类型或某些目标基因座上,基本编辑效率可能会低得令人无法接受。原因之一可能是缺乏针对未编辑细胞存活的选择性压力。关于原核生物中ABE的研究很少,可能是由于TadA的编辑效率相对较低。提高编辑效率是建立基础编辑技术尤其是ABE技术的关键。在这项工作中,对非编辑单元格施加了选择性压力,以提高基本编辑效率。首先,我们将nCas9或dCas9与TadA融合在一起,以比较nCas9-TadA和dCas9-TadA融合复合物在模型原核大肠杆菌中的编辑效率。尽管nCas9-TadA能够以中等效率实现A到G的基础编辑(ABE),但dCas9-TadA的效率却非常低。为了富集编辑后的细胞并提高碱基编辑效率,我们利用了活性Cas9诱导的双链断裂,从而导致原核细胞死亡。通过在碱基编辑过程中引入具有与nCas9-TadA相同的编辑gRNA的诱导型活性Cas9,具有未编辑靶碱基的细胞仍然易受Cas9攻击并被淘汰。因此,建立了复查基础编辑(DBE)方法,从而大大提高了ABE在大肠杆菌中的编辑效率,在某些位置达到了99.0%。通过对未编辑的细胞施加选择性压力,DBE策略还可以应用于各种情况,以提高许多不同的碱基编辑靶标的效率,甚至可以用于表观遗传学DNA修饰技术。
更新日期:2019-12-03
中文翻译:

仔细检查基本编辑,以实现有效的A到G转换。
随着CRISPR / Cas9技术的发展,转换特定碱基的新一代编辑方法实现了精确的单碱基突变。迄今为止,已经分别使用胞苷脱氨酶APOBEC1和腺苷脱氨酶(TadA)实现了胞嘧啶向胸苷的转化和腺嘌呤向鸟嘌呤的转化。但是,在某些细胞类型或某些目标基因座上,基本编辑效率可能会低得令人无法接受。原因之一可能是缺乏针对未编辑细胞存活的选择性压力。关于原核生物中ABE的研究很少,可能是由于TadA的编辑效率相对较低。提高编辑效率是建立基础编辑技术尤其是ABE技术的关键。在这项工作中,对非编辑单元格施加了选择性压力,以提高基本编辑效率。首先,我们将nCas9或dCas9与TadA融合在一起,以比较nCas9-TadA和dCas9-TadA融合复合物在模型原核大肠杆菌中的编辑效率。尽管nCas9-TadA能够以中等效率实现A到G的基础编辑(ABE),但dCas9-TadA的效率却非常低。为了富集编辑后的细胞并提高碱基编辑效率,我们利用了活性Cas9诱导的双链断裂,从而导致原核细胞死亡。通过在碱基编辑过程中引入具有与nCas9-TadA相同的编辑gRNA的诱导型活性Cas9,具有未编辑靶碱基的细胞仍然易受Cas9攻击并被淘汰。因此,建立了复查基础编辑(DBE)方法,从而大大提高了ABE在大肠杆菌中的编辑效率,在某些位置达到了99.0%。通过对未编辑的细胞施加选择性压力,DBE策略还可以应用于各种情况,以提高许多不同的碱基编辑靶标的效率,甚至可以用于表观遗传学DNA修饰技术。


























 京公网安备 11010802027423号
京公网安备 11010802027423号