当前位置:
X-MOL 学术
›
Nat. Methods
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Time-resolved imaging-based CRISPRi screening.
Nature Methods ( IF 48.0 ) Pub Date : 2019-11-18 , DOI: 10.1038/s41592-019-0629-y Daniel Camsund 1 , Michael J Lawson 1, 2 , Jimmy Larsson 1 , Daniel Jones 1 , Spartak Zikrin 1 , David Fange 1 , Johan Elf 1
Nature Methods ( IF 48.0 ) Pub Date : 2019-11-18 , DOI: 10.1038/s41592-019-0629-y Daniel Camsund 1 , Michael J Lawson 1, 2 , Jimmy Larsson 1 , Daniel Jones 1 , Spartak Zikrin 1 , David Fange 1 , Johan Elf 1
Affiliation
|
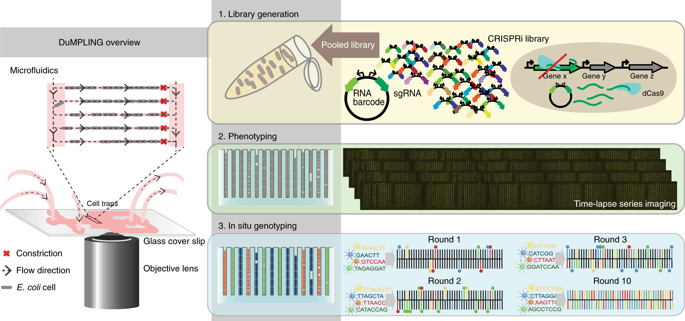
Our ability to connect genotypic variation to biologically important phenotypes has been seriously limited by the gap between live-cell microscopy and library-scale genomic engineering. Here, we show how in situ genotyping of a library of strains after time-lapse imaging in a microfluidic device overcomes this problem. We determine how 235 different CRISPR interference knockdowns impact the coordination of the replication and division cycles of Escherichia coli by monitoring the location of replication forks throughout on average >500 cell cycles per knockdown. Subsequent in situ genotyping allows us to map each phenotype distribution to a specific genetic perturbation to determine which genes are important for cell cycle control. The single-cell time-resolved assay allows us to determine the distribution of single-cell growth rates, cell division sizes and replication initiation volumes. The technology presented in this study enables genome-scale screens of most live-cell microscopy assays.
中文翻译:

基于时间分辨成像的 CRISPRi 筛查。
我们将基因型变异与生物学上重要的表型联系起来的能力受到活细胞显微镜和图书馆规模基因组工程之间差距的严重限制。在这里,我们展示了如何在微流体设备中进行延时成像后对菌株库进行原位基因分型来克服这个问题。我们通过监测复制叉的位置来确定 235 种不同的 CRISPR 干扰击倒如何影响大肠杆菌复制和分裂周期的协调,每次击倒平均 >500 个细胞周期。随后的原位基因分型使我们能够将每个表型分布映射到特定的遗传扰动,以确定哪些基因对细胞周期控制很重要。单细胞时间分辨分析使我们能够确定单细胞生长速率的分布,细胞分裂大小和复制起始体积。本研究中介绍的技术使大多数活细胞显微镜检测的基因组规模筛选成为可能。
更新日期:2019-11-18
中文翻译:

基于时间分辨成像的 CRISPRi 筛查。
我们将基因型变异与生物学上重要的表型联系起来的能力受到活细胞显微镜和图书馆规模基因组工程之间差距的严重限制。在这里,我们展示了如何在微流体设备中进行延时成像后对菌株库进行原位基因分型来克服这个问题。我们通过监测复制叉的位置来确定 235 种不同的 CRISPR 干扰击倒如何影响大肠杆菌复制和分裂周期的协调,每次击倒平均 >500 个细胞周期。随后的原位基因分型使我们能够将每个表型分布映射到特定的遗传扰动,以确定哪些基因对细胞周期控制很重要。单细胞时间分辨分析使我们能够确定单细胞生长速率的分布,细胞分裂大小和复制起始体积。本研究中介绍的技术使大多数活细胞显微镜检测的基因组规模筛选成为可能。



























 京公网安备 11010802027423号
京公网安备 11010802027423号