当前位置:
X-MOL 学术
›
Nat. Protoc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Native internally calibrated chromatin immunoprecipitation for quantitative studies of histone post-translational modifications.
Nature Protocols ( IF 14.8 ) Pub Date : 2019-11-13 , DOI: 10.1038/s41596-019-0218-7 Adrian T Grzybowski 1 , Rohan N Shah 1, 2 , William F Richter 1 , Alexander J Ruthenburg 1, 3
Nature Protocols ( IF 14.8 ) Pub Date : 2019-11-13 , DOI: 10.1038/s41596-019-0218-7 Adrian T Grzybowski 1 , Rohan N Shah 1, 2 , William F Richter 1 , Alexander J Ruthenburg 1, 3
Affiliation
|
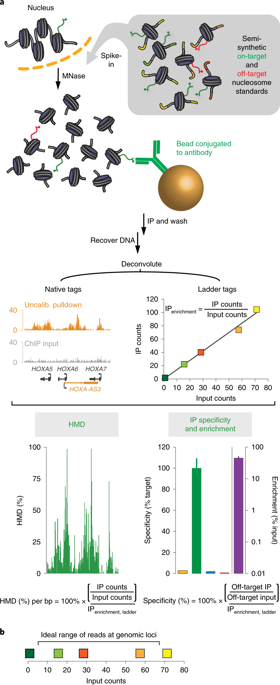
Chromatin immunoprecipitation coupled to next-generation sequencing (ChIP-seq) has served as the central method for the study of histone modifications for the past decade. In ChIP-seq analyses, antibodies selectively capture nucleosomes bearing a modification of interest and the associated DNA is then mapped to the genome to determine the distribution of the mark. This approach has several important drawbacks: (i) ChIP interpretation necessitates the assumption of perfect antibody specificity, despite growing evidence that this is often not the case. (ii) Common methods for evaluating antibody specificity in other formats have little or no bearing on specificity within a ChIP experiment. (iii) Uncalibrated ChIP is reported as relative enrichment, which is biologically meaningless outside the experimental reference frame defined by a discrete immunoprecipitation (IP), thus preventing facile comparison across experimental conditions or modifications. (iv) Differential library amplification and loading onto next-generation sequencers, as well as computational normalization, can further compromise quantitative relationships that may exist between samples. Consequently, the researcher is presented with a series of potential pitfalls and is blind to nearly all of them. Here we provide a detailed protocol for internally calibrated ChIP (ICeChIP), a method we recently developed to resolve these problems by spike-in of defined nucleosomal standards within a ChIP procedure. This protocol is optimized for specificity and quantitative power, allowing for measurement of antibody specificity and absolute measurement of histone modification density (HMD) at genomic loci on a biologically meaningful scale enabling unambiguous comparisons. We provide guidance on optimal conditions for next-generation sequencing (NGS) and instructions for data analysis. This protocol takes between 17 and 18 h, excluding time for sequencing or bioinformatic analysis. The ICeChIP procedure enables accurate measurement of histone post-translational modifications (PTMs) genome-wide in mammalian cells as well as Drosophila melanogaster and Caenorhabditis elegans, indicating suitability for use in eukaryotic cells more broadly.
中文翻译:

用于组蛋白翻译后修饰的定量研究的天然内部校准染色质免疫沉淀。
染色质免疫沉淀与二代测序 (ChIP-seq) 相结合已成为过去十年研究组蛋白修饰的中心方法。在 ChIP-seq 分析中,抗体选择性地捕获带有感兴趣修饰的核小体,然后将相关的 DNA 映射到基因组以确定标记的分布。这种方法有几个重要的缺点:(i)ChIP 解释需要假设完美的抗体特异性,尽管越来越多的证据表明情况通常并非如此。(ii) 以其他形式评估抗体特异性的常用方法对 ChIP 实验中的特异性几乎没有影响。(iii) 未校准的 ChIP 报告为相对富集,在由离散免疫沉淀 (IP) 定义的实验参考框架之外,这在生物学上是没有意义的,因此无法在实验条件或修改之间进行简单的比较。(iv) 差异文库扩增和加载到下一代测序仪,以及计算归一化,可能会进一步损害样本之间可能存在的定量关系。因此,研究人员面临一系列潜在的陷阱,几乎对所有这些陷阱都视而不见。在这里,我们提供了内部校准 ChIP (ICeChIP) 的详细协议,这是我们最近开发的一种方法,通过在 ChIP 程序中加入定义的核小体标准来解决这些问题。该协议针对特异性和定量能力进行了优化,允许在具有生物学意义的规模上测量基因组位点的抗体特异性和组蛋白修饰密度 (HMD) 的绝对测量,从而进行明确的比较。我们提供有关二代测序 (NGS) 最佳条件的指导和数据分析说明。该协议需要 17 到 18 小时,不包括测序或生物信息学分析的时间。ICeChIP 程序能够准确测量哺乳动物细胞以及黑腹果蝇和秀丽隐杆线虫中全基因组的组蛋白翻译后修饰 (PTM),表明适用于更广泛的真核细胞。我们提供有关二代测序 (NGS) 最佳条件的指导和数据分析说明。该协议需要 17 到 18 小时,不包括测序或生物信息学分析的时间。ICeChIP 程序能够准确测量哺乳动物细胞以及黑腹果蝇和秀丽隐杆线虫中全基因组的组蛋白翻译后修饰 (PTM),表明适用于更广泛的真核细胞。我们提供有关二代测序 (NGS) 最佳条件的指导和数据分析说明。该协议需要 17 到 18 小时,不包括测序或生物信息学分析的时间。ICeChIP 程序能够准确测量哺乳动物细胞以及黑腹果蝇和秀丽隐杆线虫中全基因组的组蛋白翻译后修饰 (PTM),表明适用于更广泛的真核细胞。
更新日期:2019-11-13
中文翻译:

用于组蛋白翻译后修饰的定量研究的天然内部校准染色质免疫沉淀。
染色质免疫沉淀与二代测序 (ChIP-seq) 相结合已成为过去十年研究组蛋白修饰的中心方法。在 ChIP-seq 分析中,抗体选择性地捕获带有感兴趣修饰的核小体,然后将相关的 DNA 映射到基因组以确定标记的分布。这种方法有几个重要的缺点:(i)ChIP 解释需要假设完美的抗体特异性,尽管越来越多的证据表明情况通常并非如此。(ii) 以其他形式评估抗体特异性的常用方法对 ChIP 实验中的特异性几乎没有影响。(iii) 未校准的 ChIP 报告为相对富集,在由离散免疫沉淀 (IP) 定义的实验参考框架之外,这在生物学上是没有意义的,因此无法在实验条件或修改之间进行简单的比较。(iv) 差异文库扩增和加载到下一代测序仪,以及计算归一化,可能会进一步损害样本之间可能存在的定量关系。因此,研究人员面临一系列潜在的陷阱,几乎对所有这些陷阱都视而不见。在这里,我们提供了内部校准 ChIP (ICeChIP) 的详细协议,这是我们最近开发的一种方法,通过在 ChIP 程序中加入定义的核小体标准来解决这些问题。该协议针对特异性和定量能力进行了优化,允许在具有生物学意义的规模上测量基因组位点的抗体特异性和组蛋白修饰密度 (HMD) 的绝对测量,从而进行明确的比较。我们提供有关二代测序 (NGS) 最佳条件的指导和数据分析说明。该协议需要 17 到 18 小时,不包括测序或生物信息学分析的时间。ICeChIP 程序能够准确测量哺乳动物细胞以及黑腹果蝇和秀丽隐杆线虫中全基因组的组蛋白翻译后修饰 (PTM),表明适用于更广泛的真核细胞。我们提供有关二代测序 (NGS) 最佳条件的指导和数据分析说明。该协议需要 17 到 18 小时,不包括测序或生物信息学分析的时间。ICeChIP 程序能够准确测量哺乳动物细胞以及黑腹果蝇和秀丽隐杆线虫中全基因组的组蛋白翻译后修饰 (PTM),表明适用于更广泛的真核细胞。我们提供有关二代测序 (NGS) 最佳条件的指导和数据分析说明。该协议需要 17 到 18 小时,不包括测序或生物信息学分析的时间。ICeChIP 程序能够准确测量哺乳动物细胞以及黑腹果蝇和秀丽隐杆线虫中全基因组的组蛋白翻译后修饰 (PTM),表明适用于更广泛的真核细胞。


























 京公网安备 11010802027423号
京公网安备 11010802027423号