Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
The Structural Basis for Specific Recognition of H3K14 Acetylation by Sth1 in the RSC Chromatin Remodeling Complex.
Structure ( IF 5.7 ) Pub Date : 2019-11-08 , DOI: 10.1016/j.str.2019.10.015 Guochao Chen 1 , Wei Li 1 , Fuxiang Yan 1 , Duo Wang 1 , Yong Chen 2
Structure ( IF 5.7 ) Pub Date : 2019-11-08 , DOI: 10.1016/j.str.2019.10.015 Guochao Chen 1 , Wei Li 1 , Fuxiang Yan 1 , Duo Wang 1 , Yong Chen 2
Affiliation
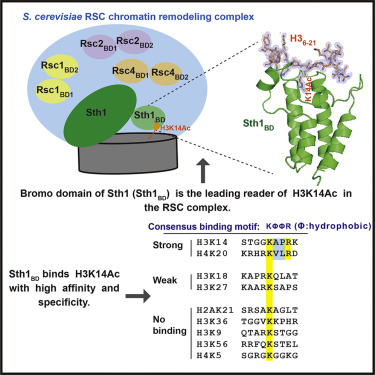
|
The Saccharomyces cerevisiae RSC (Remodel the Structure of Chromatin) complex is a chromatin-remodeling complex and plays essential roles in transcription regulation and DNA repair. The acetylation of H3 Lysine14 (H3K14Ac) enhances the RSC retention on nucleosomes and increases the remodeling activity of RSC. However, which RSC component recognizes H3K14Ac remains unclear. Here, we discovered that the bromodomain of the catalytic subunit Sth1 (Sth1BD) possessed the strongest affinity to H3K14Ac among all RSC bromodomains. The Sth1BD specifically recognized the K(Ac)ΦΦR motif (Φ stands for any hydrophobic amino acid), including H3K14Ac and H4K20Ac. We determined the crystal structures of Sth1BD at 2.40 Å resolution and Sth1BD-H3K14Ac complex at 1.40 Å resolution. The extensive interfaces between Sth1BD and H36-21 facilitate the specific and robust binding of Sth1BD to H3K14Ac. Our studies provide insights into how the RSC complex recognizes H3K14Ac to orchestrate the crosstalk between histone acetylation and chromatin remodeling.
中文翻译:

RSC染色质重塑复合物中Sth1特异性识别H3K14乙酰化的结构基础。
酿酒酵母RSC(重塑染色质结构)复合物是一种染色质重塑复合物,在转录调控和DNA修复中起重要作用。H3赖氨酸14(H3K14Ac)的乙酰化增强了RSC在核小体上的保留并增加了RSC的重塑活性。但是,尚不清楚哪个RSC组件识别H3K14Ac。在这里,我们发现催化亚基Sth1(Sth1BD)的溴结构域在所有RSC溴结构域中对H3K14Ac具有最强的亲和力。Sth1BD特别识别K(Ac)ΦΦR基序(Φ代表任何疏水性氨基酸),包括H3K14Ac和H4K20Ac。我们确定了2.40Å分辨率的Sth1BD和1.40Å分辨率的Sth1BD-H3K14Ac复合物的晶体结构。Sth1BD和H36-21之间的广泛接口促进了Sth1BD与H3K14Ac的特异性和牢固结合。我们的研究提供了有关RSC复合物如何识别H3K14Ac以协调组蛋白乙酰化和染色质重塑之间的串扰的见解。
更新日期:2019-11-08
中文翻译:

RSC染色质重塑复合物中Sth1特异性识别H3K14乙酰化的结构基础。
酿酒酵母RSC(重塑染色质结构)复合物是一种染色质重塑复合物,在转录调控和DNA修复中起重要作用。H3赖氨酸14(H3K14Ac)的乙酰化增强了RSC在核小体上的保留并增加了RSC的重塑活性。但是,尚不清楚哪个RSC组件识别H3K14Ac。在这里,我们发现催化亚基Sth1(Sth1BD)的溴结构域在所有RSC溴结构域中对H3K14Ac具有最强的亲和力。Sth1BD特别识别K(Ac)ΦΦR基序(Φ代表任何疏水性氨基酸),包括H3K14Ac和H4K20Ac。我们确定了2.40Å分辨率的Sth1BD和1.40Å分辨率的Sth1BD-H3K14Ac复合物的晶体结构。Sth1BD和H36-21之间的广泛接口促进了Sth1BD与H3K14Ac的特异性和牢固结合。我们的研究提供了有关RSC复合物如何识别H3K14Ac以协调组蛋白乙酰化和染色质重塑之间的串扰的见解。



























 京公网安备 11010802027423号
京公网安备 11010802027423号