当前位置:
X-MOL 学术
›
Mol. Phylogenet. Evol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
New insights from RADseq data on differentiation in the Hottentot golden mole species complex from South Africa.
Molecular Phylogenetics and Evolution ( IF 4.1 ) Pub Date : 2019-10-30 , DOI: 10.1016/j.ympev.2019.106667 Samantha Mynhardt 1 , Nigel C Bennett 2 , Paulette Bloomer 3
Molecular Phylogenetics and Evolution ( IF 4.1 ) Pub Date : 2019-10-30 , DOI: 10.1016/j.ympev.2019.106667 Samantha Mynhardt 1 , Nigel C Bennett 2 , Paulette Bloomer 3
Affiliation
|
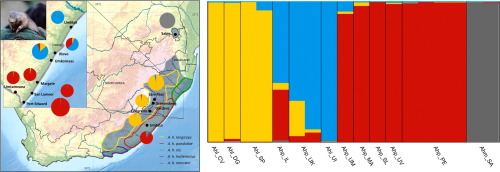
Golden moles (Family Chrysochloridae) are small subterranean mammals, endemic to sub-Saharan Africa, and many of the 21 species are listed as threatened on the IUCN Red List. Most species have highly restricted ranges; however two species, the Hottentot golden mole (Amblysomus hottentotus) and the Cape golden mole (Chrysochloris asiatica) have relatively wide ranges. We recently uncovered cryptic diversity within A. hottentotus, through a phylogeographic analysis of this taxon using two mitochondrial gene regions and a nuclear intron. To further investigate this cryptic diversity, we generated nuclear SNP data from across the genome of A. hottentotus, by means of double-digest restriction-site associated DNA sequencing (ddRADSeq), and mapped reads to the Cape golden mole genome. We conducted a phylogenetic analysis and investigated population differentiation. Our results support the distinctiveness of A. h. meesteri. Furthermore, we provide evidence from nuclear SNPs in support of our previous finding that Central coastal samples represent a unique cryptic lineage that is highly divergent from A. h. pondoliae farther south. Although mtDNA suggests that Umtata may represent a unique lineage sister to A. h. longiceps, mito-nuclear discordance from our RADseq data indicate that these samples may instead be closer to A. h. pondoliae, and therefore may not represent a distinct lineage. We stress the importance of recognizing that understudied populations, such as that of Umtata, may represent populations or ESUs under threat and in need of conservation attention. We present a high-quality filtered SNP dataset, comprising thousands of SNPs, which may serve as a useful resource for future golden mole studies. We have thus added to the growing body of research demonstrating the power and utility of RADseq to investigate population differentiation.
中文翻译:

RADseq数据对来自南非的霍滕托特金色the鼠物种群的分化产生了新的见解。
黄金mole鼠(Chrysochloridae科)是一种小型的地下哺乳动物,是撒哈拉以南非洲的特有物种,在21种物种中,有许多被列为世界自然保护联盟(IUCN)红色名录上的威胁物种。大多数物种的范围受到高度限制。然而,霍滕特金golden鼠(Amblysomus hottentotus)和开普金golden鼠(Chrysochloris asiatica)这两个物种的范围相对较大。最近,我们通过使用两个线粒体基因区域和一个核内含子对该类群进行了系统地理学分析,发现了霍滕特氏菌内的神秘多样性。为了进一步研究这种神秘的多样性,我们通过双重消化限制性位点相关的DNA测序(ddRADSeq),从嗜热链球菌的整个基因组中生成了核SNP数据,并将读段映射到金角mole基因组。我们进行了系统发育分析,并调查了种群分化。我们的结果支持A. h。的独特性。梅斯特里。此外,我们提供了来自核SNP的证据,以支持我们先前的发现,即中部沿海样品代表了一种独特的隐性谱系,与A. h有很大差异。南部更远的波陶莱亚。尽管mtDNA提示Umtata可能代表A. h。的独特血统姐妹。肱二头肌,我们RADseq数据的微核不一致表明这些样本可能更接近A. h。蓝藻科,因此可能并不代表独特的血统。我们强调必须认识到,诸如Umtata之类的被研究不足的人口可能代表处于威胁之中且需要保护的人口或ESU。我们提供了高质量的过滤SNP数据集,由数千个SNP组成,可为将来的金痣研究提供有用的资源。因此,我们增加了越来越多的研究机构,以证明RADseq在研究人口分化方面的力量和效用。
更新日期:2019-11-01
中文翻译:

RADseq数据对来自南非的霍滕托特金色the鼠物种群的分化产生了新的见解。
黄金mole鼠(Chrysochloridae科)是一种小型的地下哺乳动物,是撒哈拉以南非洲的特有物种,在21种物种中,有许多被列为世界自然保护联盟(IUCN)红色名录上的威胁物种。大多数物种的范围受到高度限制。然而,霍滕特金golden鼠(Amblysomus hottentotus)和开普金golden鼠(Chrysochloris asiatica)这两个物种的范围相对较大。最近,我们通过使用两个线粒体基因区域和一个核内含子对该类群进行了系统地理学分析,发现了霍滕特氏菌内的神秘多样性。为了进一步研究这种神秘的多样性,我们通过双重消化限制性位点相关的DNA测序(ddRADSeq),从嗜热链球菌的整个基因组中生成了核SNP数据,并将读段映射到金角mole基因组。我们进行了系统发育分析,并调查了种群分化。我们的结果支持A. h。的独特性。梅斯特里。此外,我们提供了来自核SNP的证据,以支持我们先前的发现,即中部沿海样品代表了一种独特的隐性谱系,与A. h有很大差异。南部更远的波陶莱亚。尽管mtDNA提示Umtata可能代表A. h。的独特血统姐妹。肱二头肌,我们RADseq数据的微核不一致表明这些样本可能更接近A. h。蓝藻科,因此可能并不代表独特的血统。我们强调必须认识到,诸如Umtata之类的被研究不足的人口可能代表处于威胁之中且需要保护的人口或ESU。我们提供了高质量的过滤SNP数据集,由数千个SNP组成,可为将来的金痣研究提供有用的资源。因此,我们增加了越来越多的研究机构,以证明RADseq在研究人口分化方面的力量和效用。



























 京公网安备 11010802027423号
京公网安备 11010802027423号