Cell Systems ( IF 9.3 ) Pub Date : 2019-10-30 , DOI: 10.1016/j.cels.2019.09.006 Jason A Carter 1 , Jonathan B Preall 2 , Gurinder S Atwal 2
|
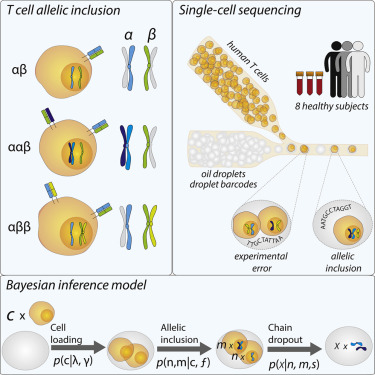
A small population of αβ T cells is characterized by the expression of more than one unique T cell receptor (TCR); this outcome is the result of “allelic inclusion,” that is, inclusion of both α- or β-chain alleles during V(D)J recombination. Limitations in single-cell sequencing technology, however, have precluded comprehensive enumeration of these dual receptor T cells. Here, we develop and experimentally validate a fully Bayesian inference model capable of reliably estimating the true rate of α and β TCR allelic inclusion across two different emulsion-barcoding single-cell sequencing platforms. We provide a database composed of over 51,000 previously unpublished allelic inclusion TCR sequence sets drawn from eight healthy individuals and show that allelic inclusion contributes a distinct and functionally important set of sequences to the human TCR repertoire. This database and a Python implementation of our statistical inference model are freely available at our Github repository (https://github.com/JasonACarter/Allelic_inclusion).
中文翻译:

人类 T 细胞受体库中等位基因包含率的贝叶斯推断。
一小群αβ T 细胞的特征是表达不止一种独特的 T 细胞受体 (TCR);这一结果是“等位基因包含”的结果,即在 V(D)J 重组过程中同时包含α - 或β - 链等位基因。然而,单细胞测序技术的局限性阻碍了对这些双受体 T 细胞的全面计数。在这里,我们开发并通过实验验证了一个完全贝叶斯推理模型,该模型能够可靠地估计α和β的真实比率跨越两个不同的乳液条形码单细胞测序平台的 TCR 等位基因包含。我们提供了一个数据库,该数据库由来自 8 个健康个体的 51,000 多个以前未发表的等位基因包含 TCR 序列集组成,并表明等位基因包含为人类 TCR 库提供了一组独特且功能重要的序列。我们的 Github 存储库 (https://github.com/JasonACarter/Allelic_inclusion) 免费提供该数据库和我们的统计推理模型的 Python 实现。


























 京公网安备 11010802027423号
京公网安备 11010802027423号