Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Predicting species abundances in a grassland biodiversity experiment: Trade‐offs between model complexity and generality
Journal of Ecology ( IF 5.5 ) Pub Date : 2019-11-19 , DOI: 10.1111/1365-2745.13316 Adam Thomas Clark 1, 2, 3 , Lindsay Ann Turnbull 4 , Andrew Tredennick 5 , Eric Allan 6 , W. Stanley Harpole 1, 2, 7 , Margaret M. Mayfield 8 , Santiago Soliveres 9 , Kathryn Barry 2, 10 , Nico Eisenhauer 2, 10 , Hans Kroon 11 , Benjamin Rosenbaum 2, 12 , Cameron Wagg 13, 14 , Alexandra Weigelt 2, 10 , Yanhao Feng 1, 2 , Christiane Roscher 1, 2 , Bernhard Schmid 14
中文翻译:

在草地生物多样性实验中预测物种丰度:模型复杂性与一般性之间的权衡
更新日期:2019-11-20
Journal of Ecology ( IF 5.5 ) Pub Date : 2019-11-19 , DOI: 10.1111/1365-2745.13316 Adam Thomas Clark 1, 2, 3 , Lindsay Ann Turnbull 4 , Andrew Tredennick 5 , Eric Allan 6 , W. Stanley Harpole 1, 2, 7 , Margaret M. Mayfield 8 , Santiago Soliveres 9 , Kathryn Barry 2, 10 , Nico Eisenhauer 2, 10 , Hans Kroon 11 , Benjamin Rosenbaum 2, 12 , Cameron Wagg 13, 14 , Alexandra Weigelt 2, 10 , Yanhao Feng 1, 2 , Christiane Roscher 1, 2 , Bernhard Schmid 14
Affiliation
|
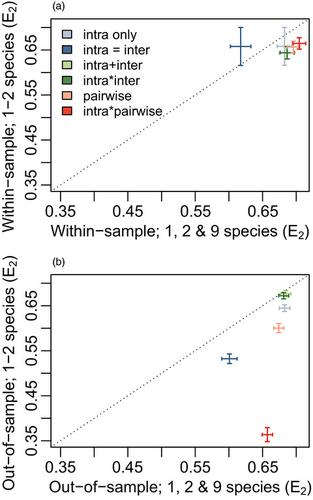
- Models of natural processes necessarily sacrifice some realism for the sake of tractability. Detailed, parameter‐rich models often provide accurate estimates of system behaviour but can be data‐hungry and difficult to operationalize. Moreover, complexity increases the danger of ‘over‐fitting’, which leads to poor performance when models are applied to novel conditions. This challenge is typically described in terms of a trade‐off between bias and variance (i.e. low accuracy vs. low precision).
- In studies of ecological communities, this trade‐off often leads to an argument about the level of detail needed to describe interactions among species. Here, we used data from a grassland biodiversity experiment containing nine locally abundant plant species (the Jena ‘dominance experiment’) to parameterize models representing six increasingly complex hypotheses about interactions. For each model, we calculated goodness‐of‐fit across different subsets of the data based on sown species richness levels, and tested how performance changed depending on whether or not the same data were used to parameterize and test the model (i.e. within vs. out‐of‐sample), and whether the range of diversity treatments being predicted fell inside or outside of the range used for parameterization.
- As expected, goodness‐of‐fit improved as a function of model complexity for all within‐sample tests. In contrast, the best out‐of‐sample performance generally resulted from models of intermediate complexity (i.e. with only two interaction coefficients per species—an intraspecific effect and a single pooled interspecific effect), especially for predictions that fell outside the range of diversity treatments used for parameterization. In accordance with other studies, our results also demonstrate that commonly used selection methods based on AIC of models fitted to the full dataset correspond more closely to within‐sample than out‐of‐sample performance.
- Synthesis. Our results demonstrate that models which include only general intra and interspecific interaction coefficients can be sufficient for estimating species‐level abundances across a wide range of contexts and may provide better out‐of‐sample performance than do more complex models. These findings serve as a reminder that simpler models may often provide a better trade‐off between bias and variance in ecological systems, particularly when applying models beyond the conditions used to parameterize them.
中文翻译:

在草地生物多样性实验中预测物种丰度:模型复杂性与一般性之间的权衡
- 为了易于处理,自然过程的模型必然会牺牲一些现实性。详细的,参数丰富的模型通常可以提供对系统行为的准确估计,但是可能需要大量数据并且难以操作。此外,复杂性增加了“过度拟合”的危险,当将模型应用于新颖条件时,这会导致性能不佳。通常以偏差和方差(即低精度与低精度)之间的权衡取舍来描述这一挑战。
- 在生态群落研究中,这种权衡常常导致人们对描述物种间相互作用所需的细节水平产生争议。在这里,我们使用了包含9种本地丰富植物物种的草原生物多样性实验(耶拿“优势实验”)的数据,对代表六个日益复杂的相互作用假设的模型进行参数化。对于每个模型,我们根据播种的物种丰富度水平计算了数据的不同子集的拟合优度,并测试了性能如何变化,这取决于是否使用相同的数据对模型进行参数化和测试(即在内部与内部)。样本外),以及预测的多样性处理范围是在参数化范围之内还是之外。
- 正如预期的那样,所有样本内测试的拟合优度都随着模型复杂度的提高而提高。相比之下,最佳的样本外性能通常是由中等复杂性模型(即每个物种只有两个相互作用系数-种内效应和单一集合种间效应)产生的,尤其是对于超出多样性处理范围的预测而言用于参数化。根据其他研究,我们的结果还表明,基于对整个数据集拟合的模型的AIC的常用选择方法与样本内性能比样本外性能更接近。
- 合成。我们的结果表明,仅包含一般种内和种间相互作用系数的模型就足以在各种情况下估计物种水平的丰度,并且比更复杂的模型可以提供更好的样本外性能。这些发现提醒我们,简单的模型通常可以在生态系统的偏差和方差之间提供更好的折衷,特别是在应用超出参数化条件的模型时。



























 京公网安备 11010802027423号
京公网安备 11010802027423号