当前位置:
X-MOL 学术
›
Org. Biomol. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Head-to-head comparison of LNA, MPγPNA, INA and Invader probes targeting mixed-sequence double-stranded DNA.
Organic & Biomolecular Chemistry ( IF 3.2 ) Pub Date : 2019-12-18 , DOI: 10.1039/c9ob02111f Raymond G Emehiser 1 , Eric Hall , Dale C Guenther , Saswata Karmakar , Patrick J Hrdlicka
Organic & Biomolecular Chemistry ( IF 3.2 ) Pub Date : 2019-12-18 , DOI: 10.1039/c9ob02111f Raymond G Emehiser 1 , Eric Hall , Dale C Guenther , Saswata Karmakar , Patrick J Hrdlicka
Affiliation
|
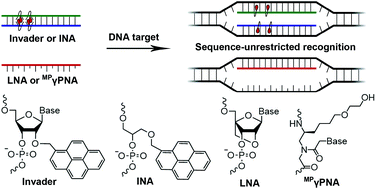
Four probe chemistries are characterized and compared with respect to thermal denaturation temperatures (Tms), thermodynamic parameters associated with duplex formation, and recognition of mixed-sequence double-stranded (ds) DNA targets: (i) oligodeoxyribonucleotides (ONs) modified with Locked Nucleic Acid (LNA) monomers, (ii) MPγPNAs, i.e., single-stranded peptide nucleic acid (PNA) probes that are functionalized at the γ-position with (R)-diethylene glycol (mini-PEG, MP) moieties, (iii) Invader probes, i.e., DNA duplexes modified with +1 interstrand zipper arrangements of 2'-O-(pyren-1-yl)methyl-RNA monomers, and (iv) intercalating nucleic acids (INAs), i.e., DNA duplexes with opposing insertions of 1-O-(1-pyrenylmethyl)glycerol bulges. Invader and INA probes, which are designed to violate the nearest-neighbor exclusion principle, denature readily, whereas the individual probe strands display exceptionally high affinity towards complementary DNA (cDNA) as indicated by increases in Tms of up to 8 °C per modification. Optimized Invader and INA probes enable efficient and highly specific recognition of mixed-sequence dsDNA targets with self-complementary regions (C50 = 30-50 nM), whereas recognition is less efficient with LNA-modified ONs and fully modified MPγPNAs due to lower cDNA affinity (LNA) and a proclivity for dimerization (LNA and MPγPNA). A Cy3-labeled Invader probe is shown to stain telomeric DNA of individual chromosomes in metaphasic spreads under non-denaturing conditions with excellent specificity.
中文翻译:

靶向混合序列双链DNA的LNA,MPγPNA,INA和Invader探针的头对头比较。
对四种探针化学特性进行了表征,并就热变性温度(Tms),与双链体形成相关的热力学参数以及识别混合序列双链(ds)DNA靶标进行了比较:(i)用锁核酸修饰的寡脱氧核糖核苷酸(ONs)酸(LNA)单体,(ii)MPγPNA,即在(γ)位上被(R)-二甘醇(mini-PEG,MP)部分官能化的单链肽核酸(PNA)探针,(iii)入侵者探针,即用2'-O-(pyren-1-yl)甲基-RNA单体的+1链间拉链修饰的DNA双链体,以及(iv)嵌入核酸(INA),即具有相反插入的DNA双链体1-O-(1-苯甲基甲基)甘油凸起。Invader和INA探针的设计违反了最近邻居排除原则,易变性,而单个探针链对互补DNA(cDNA)则显示出极高的亲和力,如每次修饰的Tms升高最高达8°C所示。经过优化的Invader和INA探针可对具有自我互补区域(C50 = 30-50 nM)的混合序列dsDNA目标进行高效,高度特异性的识别,而由于LNA修饰的ON和完全修饰的MPγPNA的识别效率较低,这是因为其cDNA亲和力较低(LNA)和二聚化倾向(LNA和MPγPNA)。已显示Cy3标记的Invader探针可在非变性条件下以优异的特异性染色变相传播中单个染色体的端粒DNA。而每个探针链对互补DNA(cDNA)则显示出极高的亲和力,如每次修饰的Tms升高高达8°C所表明的。优化的Invader和INA探针可对具有自我互补区域(C50 = 30-50 nM)的混合序列dsDNA目标进行高效,高度特异性的识别,而由于LNA修饰的ON和完全修饰的MPγPNA的识别效率较低,这是因为其cDNA亲和力较低(LNA)和二聚化倾向(LNA和MPγPNA)。已显示Cy3标记的Invader探针可在非变性条件下以优异的特异性染色变相传播中单个染色体的端粒DNA。而每个探针链对互补DNA(cDNA)则显示出极高的亲和力,如每次修饰的Tms升高高达8°C所表明的。优化的Invader和INA探针可对具有自我互补区域(C50 = 30-50 nM)的混合序列dsDNA目标进行高效,高度特异性的识别,而由于LNA修饰的ON和完全修饰的MPγPNA的识别效率较低,这是因为其cDNA亲和力较低(LNA)和二聚化倾向(LNA和MPγPNA)。已显示Cy3标记的Invader探针可在非变性条件下以优异的特异性染色变相传播中单个染色体的端粒DNA。而由于LNA修饰的ON和完全修饰的MPγPNA的识别效率较低,这是由于较低的cDNA亲和力(LNA)和二聚化的倾向(LNA和MPγPNA)。已显示Cy3标记的Invader探针可在非变性条件下以优异的特异性染色变相传播中单个染色体的端粒DNA。而由于LNA修饰的ON和完全修饰的MPγPNA的识别效率较低,这是由于较低的cDNA亲和力(LNA)和二聚化的倾向(LNA和MPγPNA)。已显示Cy3标记的Invader探针可在非变性条件下以优异的特异性染色变相传播中单个染色体的端粒DNA。
更新日期:2020-01-09
中文翻译:

靶向混合序列双链DNA的LNA,MPγPNA,INA和Invader探针的头对头比较。
对四种探针化学特性进行了表征,并就热变性温度(Tms),与双链体形成相关的热力学参数以及识别混合序列双链(ds)DNA靶标进行了比较:(i)用锁核酸修饰的寡脱氧核糖核苷酸(ONs)酸(LNA)单体,(ii)MPγPNA,即在(γ)位上被(R)-二甘醇(mini-PEG,MP)部分官能化的单链肽核酸(PNA)探针,(iii)入侵者探针,即用2'-O-(pyren-1-yl)甲基-RNA单体的+1链间拉链修饰的DNA双链体,以及(iv)嵌入核酸(INA),即具有相反插入的DNA双链体1-O-(1-苯甲基甲基)甘油凸起。Invader和INA探针的设计违反了最近邻居排除原则,易变性,而单个探针链对互补DNA(cDNA)则显示出极高的亲和力,如每次修饰的Tms升高最高达8°C所示。经过优化的Invader和INA探针可对具有自我互补区域(C50 = 30-50 nM)的混合序列dsDNA目标进行高效,高度特异性的识别,而由于LNA修饰的ON和完全修饰的MPγPNA的识别效率较低,这是因为其cDNA亲和力较低(LNA)和二聚化倾向(LNA和MPγPNA)。已显示Cy3标记的Invader探针可在非变性条件下以优异的特异性染色变相传播中单个染色体的端粒DNA。而每个探针链对互补DNA(cDNA)则显示出极高的亲和力,如每次修饰的Tms升高高达8°C所表明的。优化的Invader和INA探针可对具有自我互补区域(C50 = 30-50 nM)的混合序列dsDNA目标进行高效,高度特异性的识别,而由于LNA修饰的ON和完全修饰的MPγPNA的识别效率较低,这是因为其cDNA亲和力较低(LNA)和二聚化倾向(LNA和MPγPNA)。已显示Cy3标记的Invader探针可在非变性条件下以优异的特异性染色变相传播中单个染色体的端粒DNA。而每个探针链对互补DNA(cDNA)则显示出极高的亲和力,如每次修饰的Tms升高高达8°C所表明的。优化的Invader和INA探针可对具有自我互补区域(C50 = 30-50 nM)的混合序列dsDNA目标进行高效,高度特异性的识别,而由于LNA修饰的ON和完全修饰的MPγPNA的识别效率较低,这是因为其cDNA亲和力较低(LNA)和二聚化倾向(LNA和MPγPNA)。已显示Cy3标记的Invader探针可在非变性条件下以优异的特异性染色变相传播中单个染色体的端粒DNA。而由于LNA修饰的ON和完全修饰的MPγPNA的识别效率较低,这是由于较低的cDNA亲和力(LNA)和二聚化的倾向(LNA和MPγPNA)。已显示Cy3标记的Invader探针可在非变性条件下以优异的特异性染色变相传播中单个染色体的端粒DNA。而由于LNA修饰的ON和完全修饰的MPγPNA的识别效率较低,这是由于较低的cDNA亲和力(LNA)和二聚化的倾向(LNA和MPγPNA)。已显示Cy3标记的Invader探针可在非变性条件下以优异的特异性染色变相传播中单个染色体的端粒DNA。


























 京公网安备 11010802027423号
京公网安备 11010802027423号