当前位置:
X-MOL 学术
›
Chem. Bio. Drug Des.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
The activity prediction of indole inhibitors against HCV NS5B polymerase.
Chemical Biology & Drug Design ( IF 3 ) Pub Date : 2019-11-13 , DOI: 10.1111/cbdd.13637 Jules Muhire 1 , Hong Lin Zhai 1 , Shao Hua Lu 1 , Sha Sha Li 1 , Bo Yin 1 , Jia Ying Mi 1
Chemical Biology & Drug Design ( IF 3 ) Pub Date : 2019-11-13 , DOI: 10.1111/cbdd.13637 Jules Muhire 1 , Hong Lin Zhai 1 , Shao Hua Lu 1 , Sha Sha Li 1 , Bo Yin 1 , Jia Ying Mi 1
Affiliation
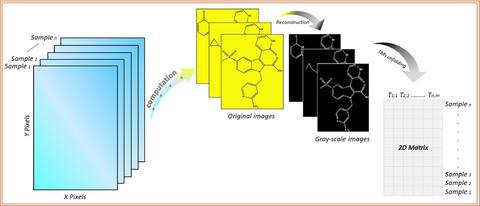
|
Non-structural viral protein 5B (NS5B) is a viral protein in hepatitis C virus. Although various inhibitors against NS5B have been found, the activity prediction of similar untested inhibitors is still highly desirable. In this respect, the Tchebichef moments (TMs) calculated from the images of molecular structures were regarded as the independent variables while the inhibitory activity (pIC50 ) was the dependent variable, and the predictive model was established by means of stepwise regression. The R-squared of leave-one-out cross-validation (Q2 ) for the training set and the R-squared of prediction ( R p 2 ) for external independent test set were 0.919 and 0.927, respectively. The obtained model was also evaluated strictly. Compared with the multivariate curve resolution with alternating least squares (MCR-ALS) and the QSAR approaches derived from the literature, the proposed method is more accurate and reliable. This study not only provides an effective approach to predict the biological activity of RNA replication's inhibitors, but also extends the QSAR modeling technique.
中文翻译:

吲哚抑制剂对 HCV NS5B 聚合酶的活性预测。
非结构病毒蛋白 5B (NS5B) 是丙型肝炎病毒中的一种病毒蛋白。尽管已经发现了各种针对 NS5B 的抑制剂,但仍然非常需要对类似的未经测试的抑制剂进行活性预测。在这方面,以分子结构图像计算的Tchebichef矩(TMs)为自变量,抑制活性(pIC50)为因变量,采用逐步回归的方法建立预测模型。训练集的留一法交叉验证 (Q2) 的 R 平方和外部独立测试集的预测 (R p 2 ) 的 R 平方分别为 0.919 和 0.927。得到的模型也进行了严格的评估。与交替最小二乘法(MCR-ALS)的多元曲线分辨率和文献中的QSAR方法相比,该方法更加准确和可靠。该研究不仅为预测RNA复制抑制剂的生物学活性提供了一种有效的方法,而且还扩展了QSAR建模技术。
更新日期:2019-11-13
中文翻译:

吲哚抑制剂对 HCV NS5B 聚合酶的活性预测。
非结构病毒蛋白 5B (NS5B) 是丙型肝炎病毒中的一种病毒蛋白。尽管已经发现了各种针对 NS5B 的抑制剂,但仍然非常需要对类似的未经测试的抑制剂进行活性预测。在这方面,以分子结构图像计算的Tchebichef矩(TMs)为自变量,抑制活性(pIC50)为因变量,采用逐步回归的方法建立预测模型。训练集的留一法交叉验证 (Q2) 的 R 平方和外部独立测试集的预测 (R p 2 ) 的 R 平方分别为 0.919 和 0.927。得到的模型也进行了严格的评估。与交替最小二乘法(MCR-ALS)的多元曲线分辨率和文献中的QSAR方法相比,该方法更加准确和可靠。该研究不仅为预测RNA复制抑制剂的生物学活性提供了一种有效的方法,而且还扩展了QSAR建模技术。



























 京公网安备 11010802027423号
京公网安备 11010802027423号