当前位置:
X-MOL 学术
›
npj Syst. Biol. Appl.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
ModelBricks-modules for reproducible modeling improving model annotation and provenance.
npj Systems Biology and Applications ( IF 4 ) Pub Date : 2019-10-08 , DOI: 10.1038/s41540-019-0114-3 Ann E Cowan 1, 2 , Pedro Mendes 1, 3, 4 , Michael L Blinov 1, 5
npj Systems Biology and Applications ( IF 4 ) Pub Date : 2019-10-08 , DOI: 10.1038/s41540-019-0114-3 Ann E Cowan 1, 2 , Pedro Mendes 1, 3, 4 , Michael L Blinov 1, 5
Affiliation
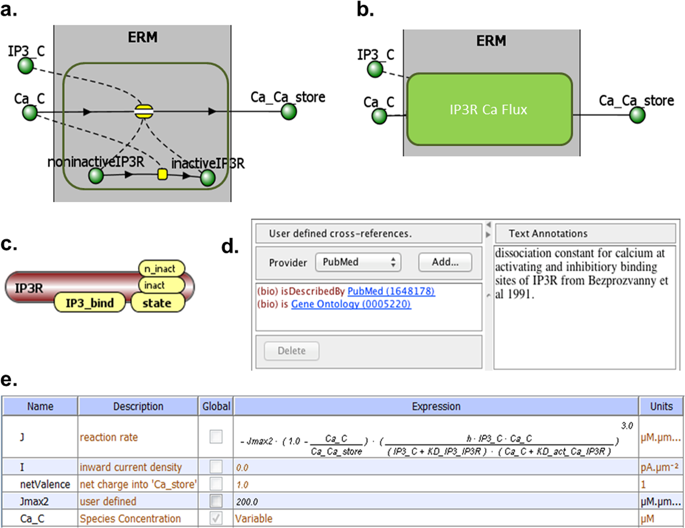
|
Most computational models in biology are built and intended for "single-use"; the lack of appropriate annotation creates models where the assumptions are unknown, and model elements are not uniquely identified. Simply recreating a simulation result from a publication can be daunting; expanding models to new and more complex situations is a herculean task. As a result, new models are almost always created anew, repeating literature searches for kinetic parameters, initial conditions and modeling specifics. It is akin to building a brick house starting with a pile of clay. Here we discuss a concept for building annotated, reusable models, by starting with small well-annotated modules we call ModelBricks. Curated ModelBricks, accessible through an open database, could be used to construct new models that will inherit ModelBricks annotations and thus be easier to understand and reuse. Key features of ModelBricks include reliance on a commonly used standard language (SBML), rule-based specification describing species as a collection of uniquely identifiable molecules, association with model specific numerical parameters, and more common annotations. Physical bricks can vary substantively; likewise, to be useful the structure of ModelBricks must be highly flexible-it should encapsulate mechanisms from single reactions to multiple reactions in a complex process. Ultimately, a modeler would be able to construct large models by using multiple ModelBricks, preserving annotations and provenance of model elements, resulting in a highly annotated model. We envision the library of ModelBricks to rapidly grow from community contributions. Persistent citable references will incentivize model creators to contribute new ModelBricks.
中文翻译:

ModelBricks - 用于可重复建模的模块,改进模型注释和来源。
大多数生物学计算模型都是为“一次性”而构建的;缺乏适当的注释会创建假设未知且模型元素无法唯一标识的模型。简单地从出版物中重新创建模拟结果可能会令人望而生畏。将模型扩展到新的、更复杂的情况是一项艰巨的任务。因此,新模型几乎总是重新创建,重复文献搜索动力学参数、初始条件和建模细节。这类似于从一堆粘土开始建造一座砖房。在这里,我们讨论构建带注释、可重用模型的概念,从我们称为 ModelBricks 的注释良好的小型模块开始。精心策划的 ModelBricks 可通过开放数据库访问,可用于构建继承 ModelBricks 注释的新模型,从而更易于理解和重用。ModelBricks 的主要功能包括依赖常用标准语言 (SBML)、基于规则的规范(将物种描述为唯一可识别分子的集合)、与模型特定数值参数的关联以及更常见的注释。物理砖块可能会有很大差异;同样,要发挥作用,ModelBricks 的结构必须高度灵活,它应该封装从单个反应到复杂过程中的多个反应的机制。最终,建模者将能够通过使用多个 ModelBrick 构建大型模型,保留模型元素的注释和来源,从而生成高度注释的模型。我们预计 ModelBricks 库将通过社区贡献快速发展。持久的可引用参考文献将激励模型创建者贡献新的 ModelBricks。
更新日期:2019-10-08
中文翻译:

ModelBricks - 用于可重复建模的模块,改进模型注释和来源。
大多数生物学计算模型都是为“一次性”而构建的;缺乏适当的注释会创建假设未知且模型元素无法唯一标识的模型。简单地从出版物中重新创建模拟结果可能会令人望而生畏。将模型扩展到新的、更复杂的情况是一项艰巨的任务。因此,新模型几乎总是重新创建,重复文献搜索动力学参数、初始条件和建模细节。这类似于从一堆粘土开始建造一座砖房。在这里,我们讨论构建带注释、可重用模型的概念,从我们称为 ModelBricks 的注释良好的小型模块开始。精心策划的 ModelBricks 可通过开放数据库访问,可用于构建继承 ModelBricks 注释的新模型,从而更易于理解和重用。ModelBricks 的主要功能包括依赖常用标准语言 (SBML)、基于规则的规范(将物种描述为唯一可识别分子的集合)、与模型特定数值参数的关联以及更常见的注释。物理砖块可能会有很大差异;同样,要发挥作用,ModelBricks 的结构必须高度灵活,它应该封装从单个反应到复杂过程中的多个反应的机制。最终,建模者将能够通过使用多个 ModelBrick 构建大型模型,保留模型元素的注释和来源,从而生成高度注释的模型。我们预计 ModelBricks 库将通过社区贡献快速发展。持久的可引用参考文献将激励模型创建者贡献新的 ModelBricks。



























 京公网安备 11010802027423号
京公网安备 11010802027423号