npj Biofilms and Microbiomes ( IF 9.2 ) Pub Date : 2019-09-19 , DOI: 10.1038/s41522-019-0099-0 Julia L. E. Willett , Michelle M. Ji , Gary M. Dunny
|
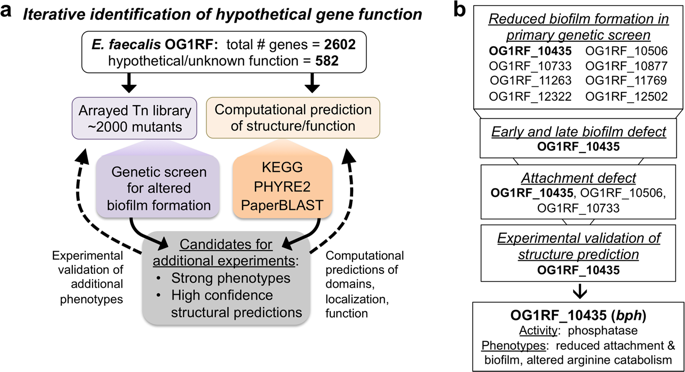
Enterococcus faecalis is a commensal organism as well as an important nosocomial pathogen, and its infections are typically linked to biofilm formation. Nearly 25% of the E. faecalis OG1RF genome encodes hypothetical genes or genes of unknown function. Elucidating their function and how these gene products influence biofilm formation is critical for understanding E. faecalis biology. To identify uncharacterized early biofilm determinants, we performed a genetic screen using an arrayed transposon (Tn) library containing ~2000 mutants in hypothetical genes/intergenic regions and identified eight uncharacterized predicted protein-coding genes required for biofilm formation. We demonstrate that OG1RF_10435 encodes a phosphatase that modulates global protein expression and arginine catabolism and propose renaming this gene bph (biofilm phosphatase). We present a workflow for combining phenotype-driven experimental and computational evaluation of hypothetical gene products in E. faecalis, which can be used to study hypothetical genes required for biofilm formation and other phenotypes of diverse bacteria.
中文翻译:

利用生物膜表型对粪肠球菌中的假设基因进行功能表征。
粪肠球菌是常见的生物,也是重要的医院病原体,其感染通常与生物膜形成有关。粪肠球菌OG1RF基因组中将近25%编码假想基因或功能未知的基因。阐明其功能以及这些基因产物如何影响生物膜形成对于理解粪肠球菌至关重要生物学。为了鉴定未表征的早期生物膜决定簇,我们使用阵列转座子(Tn)文库进行了基因筛选,该库在假设的基因/基因间区域中包含〜2000个突变体,并鉴定了形成生物膜所需的八个未表征的预测蛋白质编码基因。我们证明OG1RF_10435编码一种磷酸酶,可调节全局蛋白表达和精氨酸分解代谢,并提议重命名该基因bph(生物膜磷酸酶)。我们提出了一种工作流程,用于结合粪肠球菌中假设基因产物的表型驱动实验和计算评估,该工作流可用于研究生物膜形成和其他细菌其他表型所需的假设基因。



























 京公网安备 11010802027423号
京公网安备 11010802027423号