当前位置:
X-MOL 学术
›
WIREs Comput. Mol. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Bridging chromatin structure and function over a range of experimental spatial and temporal scales by molecular modeling
Wiley Interdisciplinary Reviews: Computational Molecular Science ( IF 11.4 ) Pub Date : 2019-08-06 , DOI: 10.1002/wcms.1434 Stephanie Portillo‐Ledesma 1 , Tamar Schlick 1, 2, 3
Wiley Interdisciplinary Reviews: Computational Molecular Science ( IF 11.4 ) Pub Date : 2019-08-06 , DOI: 10.1002/wcms.1434 Stephanie Portillo‐Ledesma 1 , Tamar Schlick 1, 2, 3
Affiliation
|
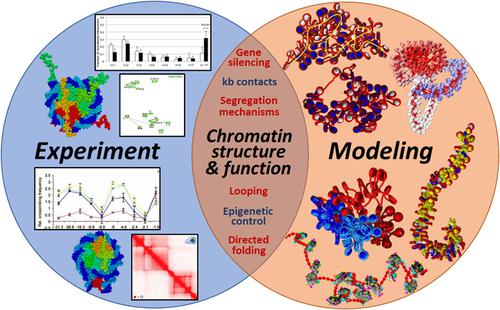
Chromatin structure, dynamics, and function are being intensely investigated by a variety of methods, including microscopy, X‐ray diffraction, nuclear magnetic resonance, biochemical crosslinking, chromosome conformation capture, and computation. The modeling helps interpret experimental data and generate configurations and mechanisms related to the three‐dimensional organization and function of the genome. Experimental contact maps, in particular, as obtained by a variety of chromosome conformation capture methods, are of increasing interest due to their implications on genome structure and regulation on many levels. In this perspective, using seven examples from our group's studies, we illustrate how molecular modeling can help interpret such experimental data. Specifically, we show how computed contact maps related to experimental systems can help interpret structures of nucleosomes, chromatin higher‐order folding, domain segregation mechanisms, gene organization, and the effect on chromatin structure of external and internal fiber parameters, such as nucleosome positioning, presence of nucleosome free regions, histone post‐translational modifications, and linker histone binding. We argue that such computations on multiple spatial and temporal scales will be increasingly important for the integration of genomic, epigenomic, and biophysical data on chromatin structure and related cellular processes.
中文翻译:

通过分子建模在一系列实验空间和时间尺度上桥接染色质结构和功能
染色质的结构,动力学和功能正在通过各种方法进行深入研究,包括显微镜检查,X射线衍射,核磁共振,生化交联,染色体构象捕获和计算。该建模有助于解释实验数据并生成与基因组的三维组织和功能有关的构型和机制。特别是通过各种染色体构象捕获方法获得的实验性接触图,由于其对基因组结构和许多水平的调控作用而引起了越来越多的关注。从这个角度出发,我们使用小组研究中的七个示例,说明了分子建模如何帮助解释此类实验数据。具体来说,我们展示了与实验系统相关的计算接触图如何帮助解释核小体的结构,染色质高阶折叠,结构域分离机制,基因组织以及对外部和内部纤维参数如核小体定位,存在核糖核酸对染色质结构的影响无核小体区域,组蛋白翻译后修饰和接头组蛋白结合。我们认为,这样的多种时空尺度的计算对于整合染色质结构和相关细胞过程的基因组,表观基因组和生物物理数据将变得越来越重要。例如核小体定位,核小体自由区的存在,组蛋白翻译后修饰和接头组蛋白结合。我们认为,这样的多种时空尺度的计算对于整合染色质结构和相关细胞过程的基因组,表观基因组和生物物理数据将变得越来越重要。例如核小体定位,核小体自由区的存在,组蛋白翻译后修饰和接头组蛋白结合。我们认为,这样的多种时空尺度的计算对于整合染色质结构和相关细胞过程的基因组,表观基因组和生物物理数据将变得越来越重要。
更新日期:2019-08-07
中文翻译:

通过分子建模在一系列实验空间和时间尺度上桥接染色质结构和功能
染色质的结构,动力学和功能正在通过各种方法进行深入研究,包括显微镜检查,X射线衍射,核磁共振,生化交联,染色体构象捕获和计算。该建模有助于解释实验数据并生成与基因组的三维组织和功能有关的构型和机制。特别是通过各种染色体构象捕获方法获得的实验性接触图,由于其对基因组结构和许多水平的调控作用而引起了越来越多的关注。从这个角度出发,我们使用小组研究中的七个示例,说明了分子建模如何帮助解释此类实验数据。具体来说,我们展示了与实验系统相关的计算接触图如何帮助解释核小体的结构,染色质高阶折叠,结构域分离机制,基因组织以及对外部和内部纤维参数如核小体定位,存在核糖核酸对染色质结构的影响无核小体区域,组蛋白翻译后修饰和接头组蛋白结合。我们认为,这样的多种时空尺度的计算对于整合染色质结构和相关细胞过程的基因组,表观基因组和生物物理数据将变得越来越重要。例如核小体定位,核小体自由区的存在,组蛋白翻译后修饰和接头组蛋白结合。我们认为,这样的多种时空尺度的计算对于整合染色质结构和相关细胞过程的基因组,表观基因组和生物物理数据将变得越来越重要。例如核小体定位,核小体自由区的存在,组蛋白翻译后修饰和接头组蛋白结合。我们认为,这样的多种时空尺度的计算对于整合染色质结构和相关细胞过程的基因组,表观基因组和生物物理数据将变得越来越重要。


























 京公网安备 11010802027423号
京公网安备 11010802027423号