当前位置:
X-MOL 学术
›
Nat. Protoc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
In situ observation of conformational dynamics and protein ligand-substrate interactions in outer-membrane proteins with DEER/PELDOR spectroscopy.
Nature Protocols ( IF 14.8 ) Pub Date : 2019-07-05 , DOI: 10.1038/s41596-019-0182-2 Benesh Joseph 1, 2 , Eva A Jaumann 2 , Arthur Sikora 3 , Katja Barth 2 , Thomas F Prisner 2 , David S Cafiso 3
Nature Protocols ( IF 14.8 ) Pub Date : 2019-07-05 , DOI: 10.1038/s41596-019-0182-2 Benesh Joseph 1, 2 , Eva A Jaumann 2 , Arthur Sikora 3 , Katja Barth 2 , Thomas F Prisner 2 , David S Cafiso 3
Affiliation
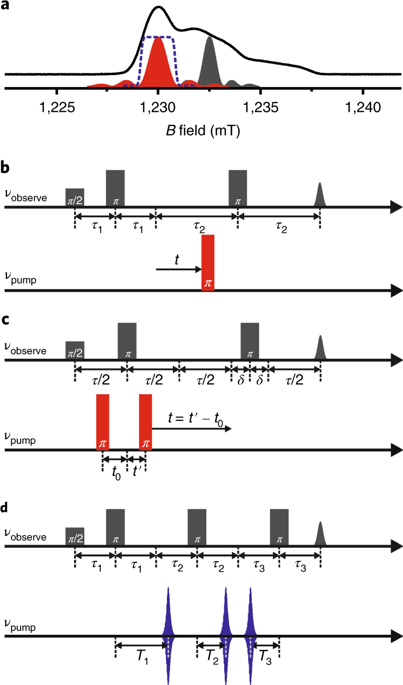
|
Observation of structure and conformational dynamics of membrane proteins at high resolution in their native environments is challenging because of the lack of suitable techniques. We have developed an approach for high-precision distance measurements in the nanometer range for outer-membrane proteins (OMPs) in intact Escherichia coli and native membranes. OMPs in Gram-negative bacteria rarely have reactive cysteines. This enables in situ labeling of engineered cysteines with a methanethiosulfonate spin label (MTSL) with minimal background signals. Following overexpression of the target protein, spin labeling is performed with E. coli or isolated outer membranes (OMs) under selective conditions. The interspin distances are measured in situ, using pulsed electron-electron double resonance (PELDOR or DEER) spectroscopy. The residual background signals, which are problematic for in situ structural biology, contribute specifically to the intermolecular part of the signal and can be selectively removed to extract the desired interspin distance distribution. The initial cloning stage can take 5-7 d, and the subsequent protein expression, OM isolation, spin labeling, PELDOR experiment, and data analysis typically take 4-5 d. The described protocol provides a general strategy for observing protein ligand-substrate interactions, oligomerization, and conformational dynamics of OMPs in their native OM and intact E. coli.
中文翻译:

用 DEER/PELDOR 光谱原位观察外膜蛋白中的构象动力学和蛋白质配体-底物相互作用。
由于缺乏合适的技术,在天然环境中以高分辨率观察膜蛋白的结构和构象动力学具有挑战性。我们开发了一种用于在纳米范围内对完整大肠杆菌和天然膜中的外膜蛋白 (OMP) 进行高精度距离测量的方法。革兰氏阴性菌中的 OMP 很少有反应性半胱氨酸。这使得使用甲硫代磺酸盐自旋标记 (MTSL) 对工程半胱氨酸进行原位标记,并具有最小的背景信号。在目标蛋白过表达后,在选择性条件下用大肠杆菌或分离的外膜 (OM) 进行自旋标记。使用脉冲电子-电子双共振(PELDOR 或 DEER)光谱原位测量自旋间距。残留的背景信号对于原位结构生物学来说是有问题的,它特别有助于信号的分子间部分,并且可以选择性地去除以提取所需的自旋间距离分布。初始克隆阶段可能需要 5-7 天,随后的蛋白质表达、OM 分离、自旋标记、PELDOR 实验和数据分析通常需要 4-5 天。所描述的协议提供了观察蛋白质配体-底物相互作用、寡聚化和 OMP 在其天然 OM 和完整大肠杆菌中的构象动力学的一般策略。随后的蛋白质表达、OM 分离、自旋标记、PELDOR 实验和数据分析通常需要 4-5 天。所描述的协议提供了观察蛋白质配体-底物相互作用、寡聚化和 OMP 在其天然 OM 和完整大肠杆菌中的构象动力学的一般策略。随后的蛋白质表达、OM 分离、自旋标记、PELDOR 实验和数据分析通常需要 4-5 天。所描述的协议提供了观察蛋白质配体-底物相互作用、寡聚化和 OMP 在其天然 OM 和完整大肠杆菌中的构象动力学的一般策略。
更新日期:2019-11-18
中文翻译:

用 DEER/PELDOR 光谱原位观察外膜蛋白中的构象动力学和蛋白质配体-底物相互作用。
由于缺乏合适的技术,在天然环境中以高分辨率观察膜蛋白的结构和构象动力学具有挑战性。我们开发了一种用于在纳米范围内对完整大肠杆菌和天然膜中的外膜蛋白 (OMP) 进行高精度距离测量的方法。革兰氏阴性菌中的 OMP 很少有反应性半胱氨酸。这使得使用甲硫代磺酸盐自旋标记 (MTSL) 对工程半胱氨酸进行原位标记,并具有最小的背景信号。在目标蛋白过表达后,在选择性条件下用大肠杆菌或分离的外膜 (OM) 进行自旋标记。使用脉冲电子-电子双共振(PELDOR 或 DEER)光谱原位测量自旋间距。残留的背景信号对于原位结构生物学来说是有问题的,它特别有助于信号的分子间部分,并且可以选择性地去除以提取所需的自旋间距离分布。初始克隆阶段可能需要 5-7 天,随后的蛋白质表达、OM 分离、自旋标记、PELDOR 实验和数据分析通常需要 4-5 天。所描述的协议提供了观察蛋白质配体-底物相互作用、寡聚化和 OMP 在其天然 OM 和完整大肠杆菌中的构象动力学的一般策略。随后的蛋白质表达、OM 分离、自旋标记、PELDOR 实验和数据分析通常需要 4-5 天。所描述的协议提供了观察蛋白质配体-底物相互作用、寡聚化和 OMP 在其天然 OM 和完整大肠杆菌中的构象动力学的一般策略。随后的蛋白质表达、OM 分离、自旋标记、PELDOR 实验和数据分析通常需要 4-5 天。所描述的协议提供了观察蛋白质配体-底物相互作用、寡聚化和 OMP 在其天然 OM 和完整大肠杆菌中的构象动力学的一般策略。


























 京公网安备 11010802027423号
京公网安备 11010802027423号