当前位置:
X-MOL 学术
›
Mol. Biosyst.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Hybrid deterministic/stochastic simulation of complex biochemical systems
Molecular BioSystems Pub Date : 2017-09-29 00:00:00 , DOI: 10.1039/c7mb00426e Paola Lecca 1, 2, 3 , Fabio Bagagiolo 1, 2, 3 , Marina Scarpa 2, 3, 4
Molecular BioSystems Pub Date : 2017-09-29 00:00:00 , DOI: 10.1039/c7mb00426e Paola Lecca 1, 2, 3 , Fabio Bagagiolo 1, 2, 3 , Marina Scarpa 2, 3, 4
Affiliation
|
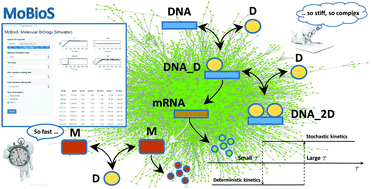
In a biological cell, cellular functions and the genetic regulatory apparatus are implemented and controlled by complex networks of chemical reactions involving genes, proteins, and enzymes. Accurate computational models are indispensable means for understanding the mechanisms behind the evolution of a complex system, not always explored with wet lab experiments. To serve their purpose, computational models, however, should be able to describe and simulate the complexity of a biological system in many of its aspects. Moreover, it should be implemented by efficient algorithms requiring the shortest possible execution time, to avoid enlarging excessively the time elapsing between data analysis and any subsequent experiment. Besides the features of their topological structure, the complexity of biological networks also refers to their dynamics, that is often non-linear and stiff. The stiffness is due to the presence of molecular species whose abundance fluctuates by many orders of magnitude. A fully stochastic simulation of a stiff system is computationally time-expensive. On the other hand, continuous models are less costly, but they fail to capture the stochastic behaviour of small populations of molecular species. We introduce a new efficient hybrid stochastic–deterministic computational model and the software tool MoBioS (MOlecular Biology Simulator) implementing it. The mathematical model of MoBioS uses continuous differential equations to describe the deterministic reactions and a Gillespie-like algorithm to describe the stochastic ones. Unlike the majority of current hybrid methods, the MoBioS algorithm divides the reactions' set into fast reactions, moderate reactions, and slow reactions and implements a hysteresis switching between the stochastic model and the deterministic model. Fast reactions are approximated as continuous-deterministic processes and modelled by deterministic rate equations. Moderate reactions are those whose reaction waiting time is greater than the fast reaction waiting time but smaller than the slow reaction waiting time. A moderate reaction is approximated as a stochastic (deterministic) process if it was classified as a stochastic (deterministic) process at the time at which it crosses the threshold of low (high) waiting time. A Gillespie First Reaction Method is implemented to select and execute the slow reactions. The performances of MoBios were tested on a typical example of hybrid dynamics: that is the DNA transcription regulation. The simulated dynamic profile of the reagents’ abundance and the estimate of the error introduced by the fully deterministic approach were used to evaluate the consistency of the computational model and that of the software tool.
中文翻译:

复杂生化系统的混合确定性/随机模拟
在生物细胞中,细胞功能和遗传调控装置是由涉及基因,蛋白质和酶的复杂化学反应网络来实现和控制的。准确的计算模型是了解复杂系统演化机理的必不可少的手段,并非总是通过湿实验室实验来探索。为了达到其目的,计算模型应该能够在生物系统的许多方面描述和模拟生物系统的复杂性。而且,应该通过需要最短执行时间的高效算法来实现,以避免过多地增加数据分析与后续实验之间的时间间隔。除了其拓扑结构的特征之外,生物网络的复杂性还涉及其动态性,通常是非线性和僵硬的。刚度归因于分子种类的丰度波动了许多数量级。刚性系统的完全随机模拟在计算上耗时。另一方面,连续模型的成本较低,但是它们无法捕获小分子物种种群的随机行为。我们介绍了一种新的高效混合随机确定性计算模型,以及实现该模型的软件工具MoBioS(分子生物学模拟器)。MoBioS的数学模型使用连续微分方程描述确定性反应,并使用类似于Gillespie的算法描述随机反应。与大多数当前的混合方法不同,MoBioS算法将反应集分为快速反应,中度反应,反应缓慢,并在随机模型和确定性模型之间实现了滞后切换。快速反应近似为连续确定性过程,并通过确定性速率方程建模。中度反应是那些反应等待时间大于快速反应等待时间但小于慢速反应等待时间的反应。如果中度反应在超过低(高)等待时间阈值时被分类为随机(确定性)过程,则该反应近似为随机(确定性)过程。吉利斯皮第一反应方法被实施以选择和执行慢反应。MoBios的性能在混合动力学的一个典型例子上进行了测试:即DNA转录调控。
更新日期:2017-11-21
中文翻译:

复杂生化系统的混合确定性/随机模拟
在生物细胞中,细胞功能和遗传调控装置是由涉及基因,蛋白质和酶的复杂化学反应网络来实现和控制的。准确的计算模型是了解复杂系统演化机理的必不可少的手段,并非总是通过湿实验室实验来探索。为了达到其目的,计算模型应该能够在生物系统的许多方面描述和模拟生物系统的复杂性。而且,应该通过需要最短执行时间的高效算法来实现,以避免过多地增加数据分析与后续实验之间的时间间隔。除了其拓扑结构的特征之外,生物网络的复杂性还涉及其动态性,通常是非线性和僵硬的。刚度归因于分子种类的丰度波动了许多数量级。刚性系统的完全随机模拟在计算上耗时。另一方面,连续模型的成本较低,但是它们无法捕获小分子物种种群的随机行为。我们介绍了一种新的高效混合随机确定性计算模型,以及实现该模型的软件工具MoBioS(分子生物学模拟器)。MoBioS的数学模型使用连续微分方程描述确定性反应,并使用类似于Gillespie的算法描述随机反应。与大多数当前的混合方法不同,MoBioS算法将反应集分为快速反应,中度反应,反应缓慢,并在随机模型和确定性模型之间实现了滞后切换。快速反应近似为连续确定性过程,并通过确定性速率方程建模。中度反应是那些反应等待时间大于快速反应等待时间但小于慢速反应等待时间的反应。如果中度反应在超过低(高)等待时间阈值时被分类为随机(确定性)过程,则该反应近似为随机(确定性)过程。吉利斯皮第一反应方法被实施以选择和执行慢反应。MoBios的性能在混合动力学的一个典型例子上进行了测试:即DNA转录调控。


























 京公网安备 11010802027423号
京公网安备 11010802027423号