Chemical Engineering Journal ( IF 15.1 ) Pub Date : 2023-06-01 , DOI: 10.1016/j.cej.2023.143865 Yan Xia , Lichao Sun , Zeyu Liang , Yingjie Guo , Jing Li , Dan Tang , Yi-Xin Huo , Shuyuan Guo
|
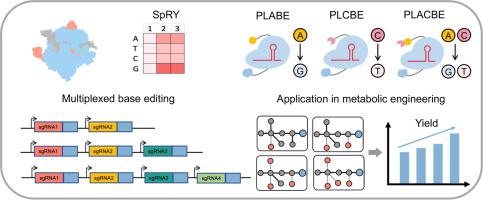
The necessity of a specific protospacer adjacent motif (PAM) greatly limited the editing scope and the application range of Cas9-dependent base editors. Here, we showed that SpRY breaks the PAM recognition barrier in Bacillus subtilis with a preference toward NRN than NYN while the PAM specificity is slightly distinct from other organisms. We developed a highly efficient PAM-less base editing toolbox for B. subtilis based on a variant of Cas9 nickase, nSpRY, yielding PAM-less adenine and/or cytosine base editors (PLABE/PLCBE/PLACBE) for A-to-G and/or C-to-T conversions. The conversion efficiency was optimized to 100% at high-activity sites and the editing window was refined to 1–2 nucleotides at pre-selected sites. Multiplexed editing for double, triple, and quadruple genes was validated simply by one cycle of editing. As a proof of concept, PLCBE was utilized to introduce premature stop codons for yielding gene knockouts and PLABE was applied to change start codons from ATG to GTG for realizing translation control. A website was designed to accelerate the inactivation of any genes of interest in B. subtilis by incorporating PAM specificity information for identifying the high-efficiency sites. Facilitated by multiplexed PAM-less editing, we successfully obtained a diversified library of strain variants for chemical production with multi-level regulations in central and competing metabolism pathways. We recruited this library to modulate metabolic fluxes in B. subtilis, achieving up to a 26.3% increase in acetoin production with a final titer of 20.8 g/L in shake-flask fermentation. By enabling in situ base editing of nearly all As and Cs in the Bacillus genome, this toolbox shows great potential in applications of metabolic engineering and codon expansion.
中文翻译:

枯草芽孢杆菌PAM-less碱基编辑工具箱的构建及其在代谢工程中的应用
特定原型间隔子相邻基序(PAM)的必要性极大地限制了 Cas9 依赖性碱基编辑器的编辑范围和应用范围。在这里,我们表明 SpRY 打破了枯草芽孢杆菌中的 PAM 识别屏障,对 NRN 的偏好高于对 NYN 的偏好,而 PAM 特异性与其他生物略有不同。我们为B开发了一个高效的无 PAM 碱基编辑工具箱。枯草芽孢杆菌属基于 Cas9 切口酶 nSpRY 的变体,产生无 PAM 的腺嘌呤和/或胞嘧啶碱基编辑器 (PLABE/PLCBE/PLACBE),用于 A-to-G 和/或 C-to-T 转换。转换效率在高活性位点优化为 100%,编辑窗口在预选位点被细化为 1-2 个核苷酸。双重、三重和四重基因的多重编辑只需一个编辑周期即可验证。作为概念证明,PLCBE 用于引入过早终止密码子以产生基因敲除,PLABE 用于将起始密码子从 ATG 更改为 GTG 以实现翻译控制。网站旨在加速枯草芽孢杆菌中任何感兴趣基因的失活通过合并 PAM 特异性信息来识别高效站点。在多重无 PAM 编辑的推动下,我们成功地获得了用于化学生产的多样化菌株变体库,在中央和竞争代谢途径中具有多级调节。我们招募了这个文库来调节枯草芽孢杆菌的代谢通量,在摇瓶发酵中实现了 26.3% 的乙偶姻产量增加,最终滴度为 20.8 g/L。通过对芽孢杆菌基因组中几乎所有的 As 和 Cs 进行原位碱基编辑,该工具箱在代谢工程和密码子扩展的应用中显示出巨大的潜力。


























 京公网安备 11010802027423号
京公网安备 11010802027423号