当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Control of Reversible Formation and Dispersion of the Three Enzyme Networks Integrating DNA Computing
Analytical Chemistry ( IF 7.4 ) Pub Date : 2023-05-30 , DOI: 10.1021/acs.analchem.3c00924 Aoi Mameuda 1 , Masahiro Takinoue 2, 3 , Koki Kamiya 1
Analytical Chemistry ( IF 7.4 ) Pub Date : 2023-05-30 , DOI: 10.1021/acs.analchem.3c00924 Aoi Mameuda 1 , Masahiro Takinoue 2, 3 , Koki Kamiya 1
Affiliation
|
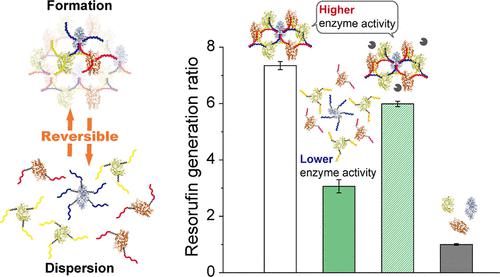
The majority of biological reactions in the cytoplasm of living cells occur via enzymatic cascade reactions. To achieve efficient enzyme cascade reactions mimicking the proximity conditions of enzymes in the cytoplasm, the proximity of each enzyme, creating a high local concentration of proteins, has been recently investigated using the conjugation of synthetic polymer molecules, proteins, and nucleic acids. Although there have been methodologies reported for the complex formation and enhanced activity of cascade reactions due to the proximity of each enzyme using DNA nanotechnology, one pair of the enzyme (GOx and HRP) complex is only assembled by the mutual independence of various shapes of the DNA structure. This study reports the network formation of three enzyme complexes assembled by a triple-branched DNA structure as a unit, thus enabling the reversible formation and dispersion of the three enzyme complex networks using single-stranded DNA, RNA, and enzymes. It was found that the activities of the three enzyme cascade reactions in the enzyme–DNA complex network were controlled by formation and dispersion of the three enzyme complex networks, due to the proximity of each enzyme with the enzyme–DNA complex network. Furthermore, three micro RNA sequences for breast cancer biomarkers were successfully detected using an enzyme–DNA complex network integrated with DNA computing. Overall, the reversible formation and dispersion of the enzyme–DNA complex network through the external stimulation of biomolecules and DNA computing provide a novel platform for controlling the production amount, diagnosis, theranostics, and biological or environmental sensing.
中文翻译:

结合DNA计算控制三种酶网络的可逆形成和分散
活细胞细胞质中的大多数生物反应通过酶促级联反应发生。为了实现模拟细胞质中酶的邻近条件的有效酶级联反应,最近使用合成聚合物分子、蛋白质和核酸的缀合研究了每种酶的邻近性,从而产生高局部浓度的蛋白质。尽管已经报道了由于使用 DNA 纳米技术使每种酶接近而导致复合物形成和级联反应活性增强的方法,但一对酶(GOx 和 HRP)复合物仅通过不同形状的相互独立来组装。 DNA 结构。这项研究报告了由三支链 DNA 结构组装成一个单元的三种酶复合物的网络形成,从而利用单链 DNA、RNA 和酶实现三种酶复合物网络的可逆形成和分散。研究发现,由于每种酶与酶-DNA 复合物网络的接近性,酶-DNA 复合物网络中三种酶级联反应的活性受到三种酶复合物网络的形成和分散的控制。此外,使用与 DNA 计算集成的酶-DNA 复合网络成功检测到了乳腺癌生物标志物的三个微 RNA 序列。总体而言,通过生物分子的外部刺激和DNA计算实现酶-DNA复合物网络的可逆形成和分散,为控制产量、诊断、治疗诊断以及生物或环境传感提供了一个新的平台。
更新日期:2023-05-30
中文翻译:

结合DNA计算控制三种酶网络的可逆形成和分散
活细胞细胞质中的大多数生物反应通过酶促级联反应发生。为了实现模拟细胞质中酶的邻近条件的有效酶级联反应,最近使用合成聚合物分子、蛋白质和核酸的缀合研究了每种酶的邻近性,从而产生高局部浓度的蛋白质。尽管已经报道了由于使用 DNA 纳米技术使每种酶接近而导致复合物形成和级联反应活性增强的方法,但一对酶(GOx 和 HRP)复合物仅通过不同形状的相互独立来组装。 DNA 结构。这项研究报告了由三支链 DNA 结构组装成一个单元的三种酶复合物的网络形成,从而利用单链 DNA、RNA 和酶实现三种酶复合物网络的可逆形成和分散。研究发现,由于每种酶与酶-DNA 复合物网络的接近性,酶-DNA 复合物网络中三种酶级联反应的活性受到三种酶复合物网络的形成和分散的控制。此外,使用与 DNA 计算集成的酶-DNA 复合网络成功检测到了乳腺癌生物标志物的三个微 RNA 序列。总体而言,通过生物分子的外部刺激和DNA计算实现酶-DNA复合物网络的可逆形成和分散,为控制产量、诊断、治疗诊断以及生物或环境传感提供了一个新的平台。



























 京公网安备 11010802027423号
京公网安备 11010802027423号