Gastroenterology ( IF 29.4 ) Pub Date : 2023-05-29 , DOI: 10.1053/j.gastro.2023.05.030 Kyung-Pil Ko 1 , Yuanjian Huang 2 , Shengzhe Zhang 1 , Gengyi Zou 1 , Bongjun Kim 1 , Jie Zhang 1 , Sohee Jun 1 , Cecilia Martin 3 , Karen J Dunbar 3 , Gizem Efe 3 , Anil K Rustgi 3 , Hiroshi Nakagawa 3 , Jae-Il Park 4
|
Background & Aims
Despite recent progress in identifying aberrant genetic and epigenetic alterations in esophageal squamous cell carcinoma (ESCC), the mechanism of ESCC initiation remains unknown.
Methods
Using CRISPR/Cas 9–based genetic ablation, we targeted 9 genes (TP53, CDKN2A, NOTCH1, NOTCH3, KMT2D, KMT2C, FAT1, FAT4, and AJUBA) in murine esophageal organoids. Transcriptomic phenotypes of organoids and chemokine released by organoids were analyzed by single-cell RNA sequencing. Tumorigenicity and immune evasion of organoids were monitored by allograft transplantation. Human ESCC single-cell RNA sequencing data sets were analyzed to classify patients and find subsets relevant to organoid models and immune evasion.
Results
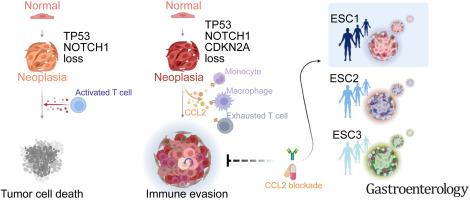
We established 32 genetically engineered esophageal organoids and identified key genetic determinants that drive ESCC initiation. A single-cell transcriptomic analysis uncovered that Trp53, Cdkn2a, and Notch1 (PCN) triple-knockout induces neoplastic features of ESCC by generating cell lineage heterogeneity and high cell plasticity. PCN knockout also generates an immunosuppressive niche enriched with exhausted T cells and M2 macrophages via the CCL2–CCR2 axis. Mechanistically, CDKN2A inactivation transactivates CCL2 via nuclear factor–κB. Moreover, comparative single-cell transcriptomic analyses stratified patients with ESCC and identified a specific subtype recapitulating the PCN-type ESCC signatures, including the high expression of CCL2 and CD274/PD-L1.
Conclusions
Our study unveils that loss of TP53, CDKN2A, and NOTCH1 induces esophageal neoplasia and immune evasion for ESCC initiation and proposes the CCL2 blockade as a viable option for targeting PCN-type ESCC.
中文翻译:

驱动食管鳞状细胞癌发生和免疫逃避的关键遗传决定因素
背景与目标
尽管最近在识别食管鳞状细胞癌 (ESCC) 的异常遗传和表观遗传改变方面取得了进展,但 ESCC 的起始机制仍然未知。
方法
使用基于 CRISPR/Cas 9 的基因消融,我们靶向小鼠食管类器官中的9 个基因( TP53、CDKN2A、NOTCH1、NOTCH3、KMT2D、KMT2C、FAT1、FAT4和AJUBA )。通过单细胞RNA测序分析类器官的转录组表型以及类器官释放的趋化因子。通过同种异体移植监测类器官的致瘤性和免疫逃避。对人类 ESCC 单细胞 RNA 测序数据集进行分析,对患者进行分类并找到与类器官模型和免疫逃避相关的子集。
结果
我们建立了 32 个基因工程食管类器官,并确定了驱动 ESCC 发生的关键遗传决定因素。单细胞转录组分析发现,Trp53、Cdkn2a和Notch1 (PCN) 三重敲除可通过产生细胞谱系异质性和高细胞可塑性来诱导食管鳞癌的肿瘤特征。PCN敲除还通过 CCL2-CCR2 轴产生富含耗尽 T 细胞和 M2 巨噬细胞的免疫抑制生态位。从机制上讲, CDKN2A失活通过核因子 -κB反式激活CCL2 。此外,比较单细胞转录组分析对 ESCC 患者进行了分层,并确定了概括 PCN 型 ESCC 特征的特定亚型,包括 CCL2 和 CD274/PD-L1 的高表达。
结论
我们的研究揭示了TP53、CDKN2A和NOTCH1的缺失会诱导食管肿瘤形成和 ESCC 起始的免疫逃避,并提出 CCL2 阻断作为针对 PCN 型 ESCC 的可行选择。



























 京公网安备 11010802027423号
京公网安备 11010802027423号