Computational and Structural Biotechnology Journal ( IF 6 ) Pub Date : 2022-12-01 , DOI: 10.1016/j.csbj.2022.11.056 Jhabindra Khanal 1 , Jeevan Kandel 2 , Hilal Tayara 3 , Kil To Chong 1, 4
|
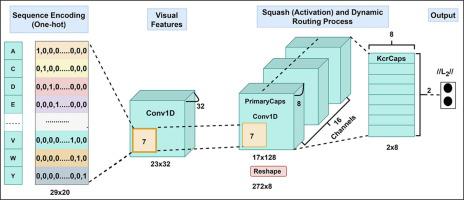
Lysine crotonylation (Kcr) is one of the most important post-translational modifications (PTMs) that is widely detected in both histone and non-histone proteins. In fact, Kcr is reported to be involved in various biological processes, such as metabolism and cell differentiation. However, the available experimental methods for Kcr site identification are laborious and costly. To effectively replace existing experimental approaches, some computational methods have been developed in the last few years. The available computational methods still lack some important aspects, as they can only identify Kcr sites on either histone-only or combined histone and nonhistone proteins. Although a tool was developed to identify Kcr sites on non-histone proteins only, its performance is inadequate and the exploration of hidden Kcr patterns (motifs) has been completely ignored, which might be significant for detailed Kcr studies. Therefore, algorithms that can more effectively predict Kcr sites on non-histone proteins with their biological meaning need to be designed. Accordingly, we developed a novel deep learning (capsule network)-based model, named CapsNh-Kcr, for Kcr site prediction, particularly focusing on non-histone proteins. Based on the independent results, the proposed model achieves an AUC of 0.9120, which is approximately 6% higher than that of previous nhKcr model in the prediction of Kcr sites on non-histone proteins. Further, we revealed, for the first time, that the proposed model can represent obvious motif distribution across Kcr sites in non-histone proteins. The source code (in Python) is publicly available at https://github.com/Jhabindra-bioinfo/CapsNh-Kcr.
中文翻译:

CapsNh-Kcr:基于胶囊网络的人非组蛋白中赖氨酸巴豆酰化位点的预测
赖氨酸巴豆酰化 (Kcr) 是最重要的翻译后修饰 (PTM) 之一,在组蛋白和非组蛋白中都被广泛检测到。事实上,据报道,Kcr 参与各种生物过程,例如新陈代谢和细胞分化。然而,用于 Kcr 位点识别的可用实验方法既费力又昂贵。为了有效地取代现有的实验方法,在过去几年中开发了一些计算方法。可用的计算方法仍然缺乏一些重要方面,因为它们只能识别仅组蛋白或组合组蛋白和非组蛋白的 Kcr 位点。尽管开发了一种工具来仅识别非组蛋白上的 Kcr 位点,它的性能不足,并且完全忽略了对隐藏的 Kcr 模式(基序)的探索,这对于详细的 Kcr 研究可能很重要。因此,需要设计能够更有效地预测非组蛋白上 Kcr 位点及其生物学意义的算法。因此,我们开发了一种新的基于深度学习(胶囊网络)的模型,名为 CapsNh-Kcr,用于 Kcr 位点预测,特别关注非组蛋白。基于独立的结果,所提出的模型在预测非组蛋白上的 Kcr 位点时实现了 0.9120 的 AUC,比之前的 nhKcr 模型高出约 6%。此外,我们首次透露,所提出的模型可以代表非组蛋白中跨 Kcr 位点的明显基序分布。 https://github.com/Jhabindra-bioinfo/CapsNh-Kcr。


























 京公网安备 11010802027423号
京公网安备 11010802027423号