当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Multisource Attention-Mechanism-Based Encoder–Decoder Model for Predicting Drug–Drug Interaction Events
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-11-30 , DOI: 10.1021/acs.jcim.2c01112 Deng Pan 1 , Lijun Quan 1, 2, 3 , Zhi Jin 1 , Taoning Chen 1 , Xuejiao Wang 1 , Jingxin Xie 1 , Tingfang Wu 1, 2, 3 , Qiang Lyu 1, 2, 3
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-11-30 , DOI: 10.1021/acs.jcim.2c01112 Deng Pan 1 , Lijun Quan 1, 2, 3 , Zhi Jin 1 , Taoning Chen 1 , Xuejiao Wang 1 , Jingxin Xie 1 , Tingfang Wu 1, 2, 3 , Qiang Lyu 1, 2, 3
Affiliation
|
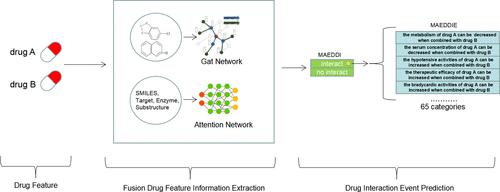
Many computational methods have been proposed to predict drug–drug interactions (DDIs), which can occur when combining drugs to treat various diseases, but most mainly utilize single-source features of drugs, which is inadequate for drug representation. To fill this gap, we propose two attention-mechanism-based encoder–decoder models that incorporate multisource information: one is MAEDDI, which can predict DDIs, and the other is MAEDDIE, which can make further DDI-associated event predictions for drug pairs with DDIs. To better express the drug feature, we used three encoding methods to encode the drugs, integrating the self-attention mechanism, cross-attention mechanism, and graph attention network to construct a multisource feature fusion network. Experiments showed that both MAEDDI and MAEDDIE performed better than some state-of-the-art methods in various validation attempts at different experimental tasks. The visualization analysis showed that the semantic features of drug pairs learned from our models had a good drug representation. In practice, MAEDDIE successfully screened 43 DDI events on favipiravir, an influenza antiviral drug, with a success rate of nearly 50%. Our model achieved competitive results, mainly owing to the design of sequence-based, structural, biochemical, and statistical multisource features. Moreover, different encoders constructed based on different features learn the interrelationship information between drug pairs, and the different representations of these drug pairs are incorporated to predict the target problem. All of these encoders were designed to better characterize the complex DDI relationships, allowing us to achieve high generalization in DDI and DDI-associated event predations.
中文翻译:

基于多源注意机制的编码器-解码器模型预测药物-药物相互作用事件
已经提出了许多计算方法来预测药物-药物相互作用 (DDI),这可能在联合药物治疗各种疾病时发生,但大多数主要利用药物的单一来源特征,这不足以用于药物表示。为了填补这一空白,我们提出了两种基于注意力机制的编码器-解码器模型,它们结合了多源信息:一种是 MAEDDI,它可以预测 DDI,另一种是 MAEDDIE,它可以对药物对进行进一步的 DDI 相关事件预测DDI。为了更好地表达药物特征,我们使用三种编码方法对药物进行编码,结合自注意力机制、交叉注意力机制和图注意力网络构建多源特征融合网络。实验表明,MAEDDI 和 MAEDDIE 在不同实验任务的各种验证尝试中都比一些最先进的方法表现更好。可视化分析表明,从我们的模型中学习到的药物对的语义特征具有良好的药物表征。在实践中,MAEDDIE成功筛选出43个流感抗病毒药物favipiravir的DDI事件,成功率接近50%。我们的模型取得了有竞争力的结果,这主要归功于基于序列、结构、生化和统计多源特征的设计。此外,基于不同特征构建的不同编码器学习药物对之间的相互关系信息,并结合这些药物对的不同表示来预测目标问题。
更新日期:2022-11-30
中文翻译:

基于多源注意机制的编码器-解码器模型预测药物-药物相互作用事件
已经提出了许多计算方法来预测药物-药物相互作用 (DDI),这可能在联合药物治疗各种疾病时发生,但大多数主要利用药物的单一来源特征,这不足以用于药物表示。为了填补这一空白,我们提出了两种基于注意力机制的编码器-解码器模型,它们结合了多源信息:一种是 MAEDDI,它可以预测 DDI,另一种是 MAEDDIE,它可以对药物对进行进一步的 DDI 相关事件预测DDI。为了更好地表达药物特征,我们使用三种编码方法对药物进行编码,结合自注意力机制、交叉注意力机制和图注意力网络构建多源特征融合网络。实验表明,MAEDDI 和 MAEDDIE 在不同实验任务的各种验证尝试中都比一些最先进的方法表现更好。可视化分析表明,从我们的模型中学习到的药物对的语义特征具有良好的药物表征。在实践中,MAEDDIE成功筛选出43个流感抗病毒药物favipiravir的DDI事件,成功率接近50%。我们的模型取得了有竞争力的结果,这主要归功于基于序列、结构、生化和统计多源特征的设计。此外,基于不同特征构建的不同编码器学习药物对之间的相互关系信息,并结合这些药物对的不同表示来预测目标问题。



























 京公网安备 11010802027423号
京公网安备 11010802027423号