Computational and Structural Biotechnology Journal ( IF 6 ) Pub Date : 2022-11-14 , DOI: 10.1016/j.csbj.2022.11.024 Chi-Chun Chen , Yu-Wei Huang , Hsuan-Cheng Huang , Wei-Cheng Lo , Ping-Chiang Lyu
|
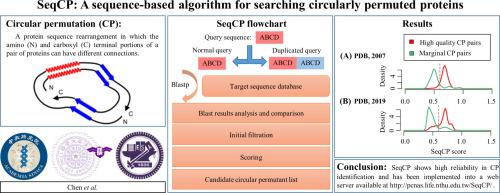
Circular permutation (CP) is a protein sequence rearrangement in which the amino- and carboxyl-termini of a protein can be created in different positions along the imaginary circularized sequence. Circularly permutated proteins usually exhibit conserved three-dimensional structures and functions. By comparing the structures of circular permutants (CPMs), protein research and bioengineering applications can be approached in ways that are difficult to achieve by traditional mutagenesis. Most current CP detection algorithms depend on structural information. Because there is a vast number of proteins with unknown structures, many CP pairs may remain unidentified. An efficient sequence-based CP detector will help identify more CP pairs and advance many protein studies. For instance, some hypothetical proteins may have CPMs with known functions and structures that are informative for functional annotation, but existing structure-based CP search methods cannot be applied when those hypothetical proteins lack structural information. Despite the considerable potential for applications, sequence-based CP search methods have not been well developed. We present a sequence-based method, SeqCP, which analyzes normal and duplicated sequence alignments to identify CPMs and determine candidate CP sites for proteins. SeqCP was trained by data obtained from the Circular Permutation Database and tested with nonredundant datasets from the Protein Data Bank. It shows high reliability in CP identification and achieves an AUC of 0.9. SeqCP has been implemented into a web server available at: http://pcnas.life.nthu.edu.tw/SeqCP/.
中文翻译:

SeqCP:一种基于序列的算法,用于搜索循环排列的蛋白质
环状排列 (CP) 是一种蛋白质序列重排,其中可以在假想的环化序列的不同位置创建蛋白质的氨基和羧基末端。循环排列的蛋白质通常表现出保守的三维结构和功能。通过比较环状置换 (CPM) 的结构,可以以传统诱变难以实现的方式进行蛋白质研究和生物工程应用。大多数当前的 CP 检测算法都依赖于结构信息。由于存在大量结构未知的蛋白质,许多 CP 对可能仍未被识别。基于序列的高效 CP 检测器将有助于识别更多 CP 对并推进许多蛋白质研究。例如,一些假设的蛋白质可能具有已知功能和结构的 CPM,这些 CPM 可为功能注释提供信息,但是当这些假设的蛋白质缺乏结构信息时,无法应用现有的基于结构的 CP 搜索方法。尽管具有相当大的应用潜力,但基于序列的 CP 搜索方法尚未得到很好的开发。我们提出了一种基于序列的方法 SeqCP,它分析正常和重复的序列比对以识别 CPM 并确定蛋白质的候选 CP 位点。SeqCP 通过从循环排列数据库获得的数据进行训练,并使用来自蛋白质数据库的非冗余数据集进行测试。它在 CP 识别中显示出高可靠性,并实现了 0.9 的 AUC。SeqCP 已在 Web 服务器中实现,网址为:http://pcnas.life.nthu.edu.tw/SeqCP/。



























 京公网安备 11010802027423号
京公网安备 11010802027423号