Journal of Advanced Research ( IF 10.7 ) Pub Date : 2022-10-18 , DOI: 10.1016/j.jare.2022.10.004 Xiao Wang 1 , Sen Wang 2 , Qiang Lin 3 , Jianjun Lu 3 , Shiyou Lv 4 , Yanxin Zhang 1 , Xuefang Wang 1 , Wei Fan 3 , Wanfei Liu 3 , Liangxiao Zhang 5 , Xiurong Zhang 1 , Jun You 1 , Peng Cui 3 , Peiwu Li 5
|
Introduction
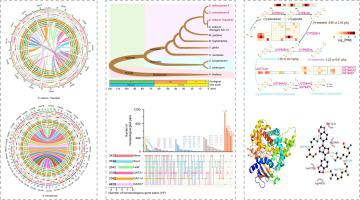
The wild tetraploid sesame (Sesamum schinzianum), an ancestral relative of diploid cultivated sesame, grows in the tropical desert of the African Plateau. As a valuable seed resource, wild sesame has several advantageous traits, such as strong environmental adaptability and an extremely high content of sesamolin in its seeds. High-quality genome assembly is essential for a detailed understanding of genome structure, genome evolution and crop improvement.
Objectives
Here, we generated two high-quality chromosome-scale genomes from S. schinzianum and a cultivated diploid elite sesame (Sesamum indicum L.) to investigate the potential genetic basis underlying these traits of wild sesame.
Methods
The long-read data from PacBio Sequel II platform and high-throughput chromosome conformation capture (Hi-C) data were used to construct high-quality sesame genome. Then dissecting the molecular mechanisms of sesame evolution and lignan biosynthesis through comparative genomics and transcriptomics.
Results
We found evidence of divergent evolution that involved differences in the number, sequence and expression level of homologous genes between the two sets of subgenomes from allotetraploids in S. schinzianum, all of which might be driven by subfunctionalization after polyploidization. Furthermore, it was found that a great number of genes involved in the stress response have undergone positive selection and resulted from gene family expansion in the wild sesame genome compared with the cultivated sesame genome, which, overall, is associated with adaptative evolution to the environment. We hypothesized that the sole functional member CYP92B14 (SscC22g35272) could be associated with high content of sesamolin in wild sesame seeds.
Conclusion
This study provides high-quality wild allotetraploid sesame and cultivated sesame genomes, reveals evolutionary features of the allotetraploid genome and provides novel insights into lignan synthesis pathways. Meanwhile, the wild sesame genome will be an important resource to conduct comparative genomic and evolutionary studies and plant improvement programmes.
中文翻译:

野生异源四倍体芝麻基因组为进化和木脂素生物合成提供了新的见解
介绍
野生四倍体芝麻(Sesamum schinzianum)是二倍体栽培芝麻的祖先亲戚,生长在非洲高原的热带沙漠中。野生芝麻作为一种宝贵的种子资源,具有环境适应性强、种子中芝麻素含量极高等优点。高质量的基因组组装对于详细了解基因组结构、基因组进化和作物改良至关重要。
目标
在这里,我们从S. schinzianum和栽培的二倍体精英芝麻(Sesamum indicum L. )中生成了两个高质量的染色体规模基因组,以研究野生芝麻这些性状背后的潜在遗传基础。
方法
利用PacBio Sequel II平台的长读长数据和高通量染色体构象捕获(Hi-C)数据构建高质量的芝麻基因组。然后通过比较基因组学和转录组学剖析芝麻进化和木脂素生物合成的分子机制。
结果
我们发现了趋异进化的证据,涉及S. schinzianum异源四倍体的两组亚基因组之间同源基因的数量、序列和表达水平的差异,所有这些都可能是由多倍体化后的亚功能化驱动的。此外,还发现野生芝麻基因组中与栽培芝麻基因组相比,大量参与应激反应的基因经历了正选择,是基因家族扩张的结果,总体上与对环境的适应性进化有关。 。我们假设唯一的功能成员 CYP92B14 (SscC22g35272) 可能与野生芝麻中高含量的芝麻素有关。
结论
本研究提供了高质量的野生异源四倍体芝麻和栽培芝麻基因组,揭示了异源四倍体基因组的进化特征,为木脂素合成途径提供了新的见解。同时,野生芝麻基因组将成为进行比较基因组和进化研究以及植物改良计划的重要资源。



























 京公网安备 11010802027423号
京公网安备 11010802027423号