Talanta ( IF 6.1 ) Pub Date : 2022-07-21 , DOI: 10.1016/j.talanta.2022.123754 Qinli Pu 1 , Yuanyuan Ye 1 , Juan Hu 1 , Cong Xie 1 , Xi Zhou 2 , Hongyan Yu 2 , Fangli Liao 1 , Song Jiang 1 , Linshan Jiang 1 , Guoming Xie 2 , Weixian Chen 1
|
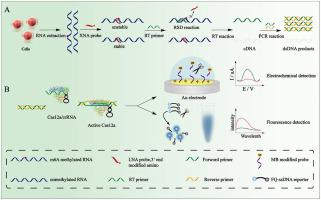
N6-methyladenosine (m6A) in RNAs is closely related to various biological progresses, but the specific regulatory mechanisms are still unclear. The existing m6A single-base resolution analysis techniques have problems of specificity and sensitivity to be improved, which can hardly meet the urgent needs of basic research and clinical applications. This work proposes a new strategy based on xeno nucleic acid (XNA) probe and CRISPR/Cas12a signal amplification for the sensitive detection of site-specific m6A modifications. According to the difference in the thermodynamic stability of hybridization between XNA probe with m6A-RNA and A-RNA, XNA was designed as a block probe to mediate m6A-RNA specific reverse transcription polymerase chain reaction (MsRT-PCR). Therefore, m6A can be specifically distinguished by converting difficult-to-test m6A modifications into easily detectable dsDNA fragments. Integration of CRISPR/Cas12a technology, skilfully designed sequences of crRNAs targeting m6A site-specific amplification dsDNA. The specificity was significantly improved through dual specific recognition of XNA probe and crRNA. Furthermore, the sensitivity of the assay was also greatly increased by the combined signal amplification of PCR and CRISPR/Cas12a. Additionally, we extend the application of CRISPR/Cas12a to flexible fluorescent and electrochemical biosensing system, which can accurately detect m6A modifications with different ranges of methylation fractions. The analysis results of m6A sites in MALAT1, ACTB and TPT1 further demonstrated the feasibility of the constructed biosensor for the accurate detection of hypomethylated samples in cells. The implementation of this work will provide strong technical support to promote the in-depth research on m6A in disease regulation mechanisms and in vitro molecular diagnosis.
中文翻译:

XNA 探针和 CRISPR/Cas12a 驱动的柔性荧光和电化学双模式生物传感器,用于灵敏检测 m6A 位点特异性 RNA 修饰
RNA中的N6-甲基腺苷(m6A)与各种生物学进程密切相关,但具体的调控机制尚不清楚。现有的m6A单碱基分辨率分析技术存在特异性和灵敏度有待提高的问题,难以满足基础研究和临床应用的迫切需求。这项工作提出了一种基于异种核酸 (XNA) 探针和 CRISPR/Cas12a 信号放大的新策略,用于灵敏检测位点特异性 m6A 修饰。根据XNA探针与m6A-RNA和A-RNA杂交热力学稳定性的差异,设计XNA作为嵌段探针介导m6A-RNA特异性逆转录聚合酶链反应(MsRT-PCR)。所以,通过将难以测试的 m6A 修饰转化为易于检测的 dsDNA 片段,可以明确区分 m6A。整合CRISPR/Cas12a技术,巧妙设计靶向m6A位点特异性扩增dsDNA的crRNA序列。通过 XNA 探针和 crRNA 的双重特异性识别,显着提高了特异性。此外,PCR 和 CRISPR/Cas12a 的联合信号放大也大大提高了检测的灵敏度。此外,我们将 CRISPR/Cas12a 的应用扩展到灵活的荧光和电化学生物传感系统,该系统可以准确检测具有不同甲基化分数范围的 m6A 修饰。MALAT1中m6A位点的分析结果,ACTB和TPT1进一步证明了构建的生物传感器用于准确检测细胞中低甲基化样品的可行性。该工作的实施将为促进m6A在疾病调控机制和体外分子诊断方面的深入研究提供强有力的技术支持。



























 京公网安备 11010802027423号
京公网安备 11010802027423号