Medical Image Analysis ( IF 10.9 ) Pub Date : 2022-07-16 , DOI: 10.1016/j.media.2022.102532 Fumin Guo 1 , Matthew Ng 2 , Grey Kuling 2 , Graham Wright 2
|
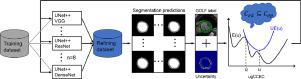
The performance of deep learning for cardiac magnetic resonance imaging (MRI) segmentation is oftentimes degraded when using small datasets and sparse annotations for training or adapting a pre-trained model to previously unseen datasets. Here, we developed and evaluated an approach to addressing some of these issues to facilitate broader use of deep learning for short-axis cardiac MRI segmentation. We developed a globally optimal label fusion (GOLF) algorithm that enforced spatial smoothness to generate consensus segmentation from segmentation predictions provided by a deep learning ensemble algorithm. The GOLF consensus was entered into an uncertainty-guided coupled continuous kernel cut (ugCCKC) algorithm that employed normalized cut, image-grid continuous regularization, and “nesting” and circular shape priors of the left ventricular myocardium (LVM) and cavity (LVC). In addition, the uncertainty measurements derived from the segmentation predictions were used to constrain the similarity of GOLF and final segmentation. We optimized ugCCKC through upper bound relaxation, for which we developed an efficient coupled continuous max-flow algorithm implemented in an iterative manner. We showed that GOLF yielded average symmetric surface distance (ASSD) 0.2–0.8 mm lower than an averaging method with higher or similar Dice similarity coefficient (DSC). We also demonstrated that ugCCKC incorporating the shape priors improved DSC by 0.01–0.05 and reduced ASSD by 0.1–0.9 mm. In addition, we integrated GOLF and ugCCKC into a deep learning ensemble algorithm by refining the segmentation of an unannotated dataset and using the refined segmentation to update the trained models. With the proposed framework, we demonstrated the capability of using relatively small datasets (5–10 subjects) with sparse (5–25% slices labeled) annotations to train a deep learning algorithm, while achieving DSC of 0.871–0.893 for LVM and 0.933–0.959 for LVC on the LVQuan dataset, and these were 0.844–0.871 for LVM and 0.923–0.931 for LVC on the ACDC dataset. Furthermore, we showed that the proposed approach can be adapted to substantially alleviate the domain shift issue. Moreover, we calculated a number of commonly used LV function measurements using the derived segmentation and observed strong correlations (Pearson –1.00, ) between algorithm and manual LV function analyses. These results suggest that the developed approaches can be used to facilitate broader application of deep learning in research and clinical cardiac MR imaging workflow.
中文翻译:

具有稀疏注释的心脏 MRI 分割:集成深度学习不确定性和形状先验
当使用小型数据集和稀疏注释进行训练或使预训练模型适应以前未见过的数据集时,心脏磁共振成像 (MRI) 分割的深度学习性能通常会下降。在这里,我们开发并评估了一种解决其中一些问题的方法,以促进更广泛地使用深度学习进行短轴心脏 MRI 分割。我们开发了一种全局最优标签融合 (GOLF) 算法,该算法强制执行空间平滑度,以根据深度学习集成算法提供的分割预测生成一致分割。GOLF 共识被纳入不确定性引导的耦合连续核切割 (ugCCKC) 算法,该算法采用归一化切割、图像网格连续正则化、以及左心室心肌 (LVM) 和腔 (LVC) 的“嵌套”和圆形先验。此外,来自分割预测的不确定性测量用于约束 GOLF 和最终分割的相似性。我们通过上限松弛优化了 ugCCKC,为此我们开发了一种以迭代方式实现的高效耦合连续最大流算法。我们表明,GOLF 产生的平均对称表面距离 (ASSD) 比具有更高或相似 Dice 相似系数 (DSC) 的平均方法低 0.2–0.8 mm。我们还证明,结合先验形状的 ugCCKC 将 DSC 提高了 0.01-0.05,并将 ASSD 降低了 0.1-0.9 mm。此外,我们通过细化未注释数据集的分割并使用细化的分割来更新训练模型,将 GOLF 和 ugCCKC 集成到深度学习集成算法中。通过提出的框架,我们展示了使用具有稀疏(5-25% 切片标记)注释的相对较小的数据集(5-10 个主题)来训练深度学习算法的能力,同时实现 LVM 的 DSC 为 0.871-0.893 和 0.933- LVQuan 数据集上的 LVC 为 0.959,LVM 为 0.844–0.871,ACDC 数据集上的 LVC 为 0.923–0.931。此外,我们表明所提出的方法可以适用于大大缓解域转移问题。此外,我们使用派生的分割和观察到的强相关性计算了许多常用的 LV 功能测量值(Pearson 我们展示了使用相对较小的数据集(5-10 个受试者)和稀疏(5-25% 切片标记)注释来训练深度学习算法的能力,同时在LVQuan 数据集,在 ACDC 数据集上,LVM 为 0.844–0.871,LVC 为 0.923–0.931。此外,我们表明所提出的方法可以适用于大大缓解域转移问题。此外,我们使用派生的分割和观察到的强相关性计算了许多常用的 LV 功能测量值(Pearson 我们展示了使用相对较小的数据集(5-10 个受试者)和稀疏(5-25% 切片标记)注释来训练深度学习算法的能力,同时在LVQuan 数据集,在 ACDC 数据集上,LVM 为 0.844–0.871,LVC 为 0.923–0.931。此外,我们表明所提出的方法可以适用于大大缓解域转移问题。此外,我们使用派生的分割和观察到的强相关性计算了许多常用的 LV 功能测量值(Pearson 在 ACDC 数据集上,LVM 为 0.844–0.871,LVC 为 0.923–0.931。此外,我们表明所提出的方法可以适用于大大缓解域转移问题。此外,我们使用派生的分割和观察到的强相关性计算了许多常用的 LV 功能测量值(Pearson 在 ACDC 数据集上,LVM 为 0.844–0.871,LVC 为 0.923–0.931。此外,我们表明所提出的方法可以适用于大大缓解域转移问题。此外,我们使用派生的分割和观察到的强相关性计算了许多常用的 LV 功能测量值(Pearson–1.00,) 在算法和手动 LV 功能分析之间。这些结果表明,所开发的方法可用于促进深度学习在研究和临床心脏 MR 成像工作流程中的更广泛应用。


























 京公网安备 11010802027423号
京公网安备 11010802027423号