Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Label-free 1D microfluidic dipstick counting of microbial colonies and bacteriophage plaques
Lab on a Chip ( IF 6.1 ) Pub Date : 2022-07-06 , DOI: 10.1039/d2lc00280a Sultan İlayda Dönmez 1 , Sarah H Needs 1 , Helen M I Osborn 1 , Nuno M Reis 2, 3 , Alexander D Edwards 1, 3
Lab on a Chip ( IF 6.1 ) Pub Date : 2022-07-06 , DOI: 10.1039/d2lc00280a Sultan İlayda Dönmez 1 , Sarah H Needs 1 , Helen M I Osborn 1 , Nuno M Reis 2, 3 , Alexander D Edwards 1, 3
Affiliation
|
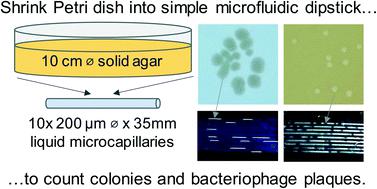
Counting viable bacterial cells and functional bacteriophage is fundamental to microbiology underpinning research, surveillance, biopharmaceuticals and diagnostics. Colony forming unit (CFU) and plaque forming unit (PFU) counting still requires slow and laborious solid culture on agar in Petri dishes or plates. Here, we show that dip-stick microfluidic strips can be used without growth indicator dye for rapid and simple CFU ml−1 and PFU ml−1 measurement. We demonstrate for the first time that fluoropolymer microcapillaries combined with digital imaging allow bacteriophage plaques to be counted rapidly in a dip-and-test format. The microfluidic length scales offer a linear 1-dimensional alternative to a 2D solid agar medium surface, with colonies or plaques clearly visible as “dashes” or “gaps”. An inexpensive open source darkfield biosensor system using Raspberry Pi imaging permits label-free detection and counting of colonies or plaques within 4–8 hours in a linear, liquid matrix within ∼200 μm inner diameter microcapillaries. We obtained full quantitative agreement between 1D microfluidic colony counting in dipsticks versus conventional 2D solid agar Petri dish plates for S. aureus and E. coli, and for T2 phage and phage K, but up to 6 times faster. Time-lapse darkfield imaging permitted detailed kinetic analysis of colony growth in the microcapillaries, providing new insight into microfluidic microbiology and colony growth, not possible with Petri dishes. Surprisingly, whilst E. coli colonies appeared earlier, subsequent colony expansion was faster along the microcapillaries for S. aureus. This may be explained by the microenvironment offered for 1D colony growth within microcapillaries, linked to a mass balance between nutrient (glucose) diffusion and bacterial growth kinetics. Counting individual colonies in liquid medium was not possible for motile strains that spread rapidly along the capillary, however inclusion of soft agar inhibited spreading, making this new simple dip-and-test counting method applicable to both motile and non-motile bacteria. Label-free dipstick colony and plaque counting has potential for many analytical microbial tasks, and the innovation of 1D colony counting has relevance to other microfluidic microbiology.
中文翻译:

微生物菌落和噬菌体斑块的无标记一维微流体试纸计数
计数活细菌细胞和功能性噬菌体是微生物学支撑研究、监测、生物制药和诊断的基础。菌落形成单位 (CFU) 和噬菌斑形成单位 (PFU) 计数仍然需要在培养皿或平板中的琼脂上进行缓慢而费力的固体培养。在这里,我们展示了在没有生长指示染料的情况下,可以使用试纸条微流控条进行快速简单的 CFU ml -1和 PFU ml -1测量。我们首次证明,含氟聚合物微毛细管与数字成像相结合,可以以浸渍和测试的形式快速计数噬菌体斑块。微流控长度标尺为 2D 固体琼脂培养基表面提供线性 1 维替代方案,菌落或斑块清晰可见为“破折号”或“间隙”。使用 Raspberry Pi 成像的廉价开源暗场生物传感器系统允许在 4-8 小时内在 200 μm 内径微毛细管内的线性液体基质中进行菌落或斑块的无标记检测和计数。我们在试纸中的 1D 微流控菌落计数与用于金黄色葡萄球菌和大肠杆菌的传统 2D 固体琼脂培养皿之间获得了完全的定量一致性,以及 T2 噬菌体和噬菌体 K,但速度快 6 倍。延时暗场成像允许对微毛细血管中的菌落生长进行详细的动力学分析,为微流体微生物学和菌落生长提供了新的见解,这在培养皿中是不可能的。令人惊讶的是,虽然大肠杆菌菌落出现得更早,但随后的菌落扩张沿着金黄色葡萄球菌的微毛细管更快. 这可以通过为微毛细管内的一维菌落生长提供的微环境来解释,这与营养(葡萄糖)扩散和细菌生长动力学之间的质量平衡有关。对于沿毛细管快速传播的运动菌株,不可能在液体培养基中计数单个菌落,但软琼脂的加入抑制了传播,使得这种新的简单的浸渍和测试计数方法适用于运动和非运动细菌。无标签试纸菌落和菌斑计数具有许多分析微生物任务的潜力,而一维菌落计数的创新与其他微流体微生物学相关。
更新日期:2022-07-06
中文翻译:

微生物菌落和噬菌体斑块的无标记一维微流体试纸计数
计数活细菌细胞和功能性噬菌体是微生物学支撑研究、监测、生物制药和诊断的基础。菌落形成单位 (CFU) 和噬菌斑形成单位 (PFU) 计数仍然需要在培养皿或平板中的琼脂上进行缓慢而费力的固体培养。在这里,我们展示了在没有生长指示染料的情况下,可以使用试纸条微流控条进行快速简单的 CFU ml -1和 PFU ml -1测量。我们首次证明,含氟聚合物微毛细管与数字成像相结合,可以以浸渍和测试的形式快速计数噬菌体斑块。微流控长度标尺为 2D 固体琼脂培养基表面提供线性 1 维替代方案,菌落或斑块清晰可见为“破折号”或“间隙”。使用 Raspberry Pi 成像的廉价开源暗场生物传感器系统允许在 4-8 小时内在 200 μm 内径微毛细管内的线性液体基质中进行菌落或斑块的无标记检测和计数。我们在试纸中的 1D 微流控菌落计数与用于金黄色葡萄球菌和大肠杆菌的传统 2D 固体琼脂培养皿之间获得了完全的定量一致性,以及 T2 噬菌体和噬菌体 K,但速度快 6 倍。延时暗场成像允许对微毛细血管中的菌落生长进行详细的动力学分析,为微流体微生物学和菌落生长提供了新的见解,这在培养皿中是不可能的。令人惊讶的是,虽然大肠杆菌菌落出现得更早,但随后的菌落扩张沿着金黄色葡萄球菌的微毛细管更快. 这可以通过为微毛细管内的一维菌落生长提供的微环境来解释,这与营养(葡萄糖)扩散和细菌生长动力学之间的质量平衡有关。对于沿毛细管快速传播的运动菌株,不可能在液体培养基中计数单个菌落,但软琼脂的加入抑制了传播,使得这种新的简单的浸渍和测试计数方法适用于运动和非运动细菌。无标签试纸菌落和菌斑计数具有许多分析微生物任务的潜力,而一维菌落计数的创新与其他微流体微生物学相关。


























 京公网安备 11010802027423号
京公网安备 11010802027423号