当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
A Novel PNGase Rc for Improved Protein N-Deglycosylation in Bioanalytics and Hydrogen–Deuterium Exchange Coupled With Mass Spectrometry Epitope Mapping under Challenging Conditions
Analytical Chemistry ( IF 7.4 ) Pub Date : 2022-06-24 , DOI: 10.1021/acs.analchem.2c01748 Marius Gramlich 1 , Sandra Maier 1 , Philipp D Kaiser 1 , Bjoern Traenkle 1 , Teresa R Wagner 1, 2 , Josef Voglmeir 3 , Dieter Stoll 1, 4 , Ulrich Rothbauer 1, 2 , Anne Zeck 1
Analytical Chemistry ( IF 7.4 ) Pub Date : 2022-06-24 , DOI: 10.1021/acs.analchem.2c01748 Marius Gramlich 1 , Sandra Maier 1 , Philipp D Kaiser 1 , Bjoern Traenkle 1 , Teresa R Wagner 1, 2 , Josef Voglmeir 3 , Dieter Stoll 1, 4 , Ulrich Rothbauer 1, 2 , Anne Zeck 1
Affiliation
|
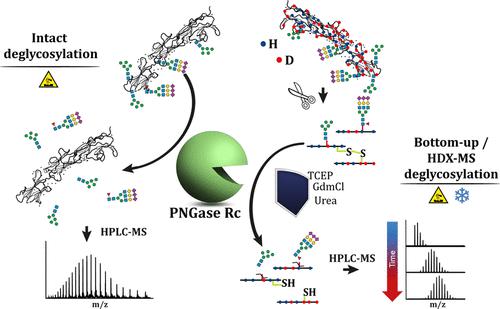
N-linked glycosylation is a ubiquitous posttranslational modification of proteins. While it plays an important role in the biological function of proteins, it often poses a major challenge for their analytical characterization. Currently available peptide N-glycanases (PNGases) are often inefficient at deglycosylating proteins due to sterically inaccessible N-glycosylation sites. This usually leads to poor sequence coverage in bottom-up analysis using liquid chromatography with tandem mass spectrometry and makes it impossible to obtain an intact mass signal in top-down MS analysis. In addition, most PNGases operate optimally only in the neutral to slightly acidic pH range and are severely compromised in the presence of reducing and denaturing substances, which limits their use for advanced bioanalysis based on hydrogen–deuterium exchange in combination with mass spectrometry (HDX-MS). Here, we present a novel peptide N-glycanase from Rudaea cellulosilytica (PNGase Rc) for which we demonstrate broad substrate specificity for N-glycan hydrolysis from multiply occupied and natively folded proteins. Our results show that PNGase Rc is functional even under challenging, HDX quenching conditions (pH 2.5, 0 °C) and in the presence of 0.4 M tris(2-carboxyethyl)phosphine, 4 M urea, and 1 M guanidinium chloride. Most importantly, we successfully applied the PNGase Rc in an HDX-MS workflow to determine the epitope of a nanobody targeting the extracellular domain of human signal-regulating protein alpha (SIRPα).
中文翻译:

一种新的 PNGase Rc,用于改进生物分析和氢-氘交换中蛋白质 N-去糖基化,并在具有挑战性的条件下结合质谱表位作图
N-连接糖基化是一种普遍存在的蛋白质翻译后修饰。虽然它在蛋白质的生物学功能中发挥着重要作用,但通常对其分析表征提出重大挑战。目前可用的肽N由于在空间上不可接近的 N-糖基化位点,β-聚糖酶 (PNGase) 通常无法有效地使蛋白质去糖基化。这通常会导致使用液相色谱和串联质谱的自下而上分析中的序列覆盖率较差,并且无法在自上而下的 MS 分析中获得完整的质量信号。此外,大多数 PNG 酶仅在中性至微酸性 pH 范围内发挥最佳作用,并且在存在还原性和变性物质时严重受损,这限制了它们在基于氢-氘交换结合质谱法的高级生物分析中的使用 (HDX-多发性硬化症)。在这里,我们提出了一种来自Rudaea cellulosilytica (PNGase Rc)的新型肽N-聚糖酶,我们证明了其广泛的底物特异性N-聚糖水解来自多重占据和天然折叠的蛋白质。我们的结果表明,即使在具有挑战性的 HDX 淬灭条件(pH 2.5, 0 °C)和存在 0.4 M 三(2-羧乙基)膦、4 M 尿素和 1 M 氯化胍的情况下,PNGase Rc 也能发挥作用。最重要的是,我们成功地将 PNGase Rc 应用于 HDX-MS 工作流程,以确定靶向人类信号调节蛋白 α (SIRPα) 细胞外结构域的纳米抗体的表位。
更新日期:2022-06-24
中文翻译:

一种新的 PNGase Rc,用于改进生物分析和氢-氘交换中蛋白质 N-去糖基化,并在具有挑战性的条件下结合质谱表位作图
N-连接糖基化是一种普遍存在的蛋白质翻译后修饰。虽然它在蛋白质的生物学功能中发挥着重要作用,但通常对其分析表征提出重大挑战。目前可用的肽N由于在空间上不可接近的 N-糖基化位点,β-聚糖酶 (PNGase) 通常无法有效地使蛋白质去糖基化。这通常会导致使用液相色谱和串联质谱的自下而上分析中的序列覆盖率较差,并且无法在自上而下的 MS 分析中获得完整的质量信号。此外,大多数 PNG 酶仅在中性至微酸性 pH 范围内发挥最佳作用,并且在存在还原性和变性物质时严重受损,这限制了它们在基于氢-氘交换结合质谱法的高级生物分析中的使用 (HDX-多发性硬化症)。在这里,我们提出了一种来自Rudaea cellulosilytica (PNGase Rc)的新型肽N-聚糖酶,我们证明了其广泛的底物特异性N-聚糖水解来自多重占据和天然折叠的蛋白质。我们的结果表明,即使在具有挑战性的 HDX 淬灭条件(pH 2.5, 0 °C)和存在 0.4 M 三(2-羧乙基)膦、4 M 尿素和 1 M 氯化胍的情况下,PNGase Rc 也能发挥作用。最重要的是,我们成功地将 PNGase Rc 应用于 HDX-MS 工作流程,以确定靶向人类信号调节蛋白 α (SIRPα) 细胞外结构域的纳米抗体的表位。



























 京公网安备 11010802027423号
京公网安备 11010802027423号