当前位置:
X-MOL 学术
›
J. Am. Chem. Soc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
The Biosynthetic Landscape of Triceptides Reveals Radical SAM Enzymes That Catalyze Cyclophane Formation on Tyr- and His-Containing Motifs
Journal of the American Chemical Society ( IF 15.0 ) Pub Date : 2022-06-21 , DOI: 10.1021/jacs.2c00521 Ryosuke Sugiyama 1 , Angelica Faith L Suarez 1 , Yohei Morishita 1 , Thi Quynh Ngoc Nguyen 1 , Yi Wei Tooh 1 , Muhammad Nur Hadi Bin Roslan 1 , Justin Lo Choy 2 , Qi Su 1 , Wei Yang Goh 1 , Gregory Adrian Gunawan 1, 3, 4 , Fong Tian Wong 3, 5 , Brandon I Morinaka 1
Journal of the American Chemical Society ( IF 15.0 ) Pub Date : 2022-06-21 , DOI: 10.1021/jacs.2c00521 Ryosuke Sugiyama 1 , Angelica Faith L Suarez 1 , Yohei Morishita 1 , Thi Quynh Ngoc Nguyen 1 , Yi Wei Tooh 1 , Muhammad Nur Hadi Bin Roslan 1 , Justin Lo Choy 2 , Qi Su 1 , Wei Yang Goh 1 , Gregory Adrian Gunawan 1, 3, 4 , Fong Tian Wong 3, 5 , Brandon I Morinaka 1
Affiliation
|
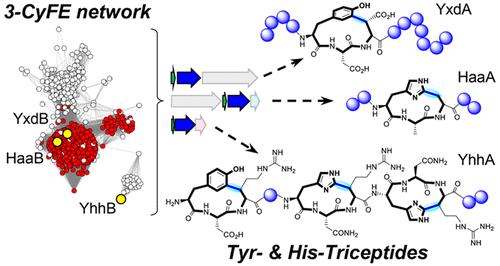
Peptide-derived cyclophanes inhabit a unique niche in the chemical space of macrocyclic peptides with several examples of pharmaceutical importance. Although both synthetic and biocatalytic methods are available for constructing these macrocycles, versatile (bio)catalysts able to incorporate a variety of amino acids that compose the macrocycle would be useful for the creation of diverse peptide cyclophanes. In this report, we synergized the use of bioinformatic tools to map the biosynthetic landscape of radical SAM enzymes (3-CyFEs) that catalyze three-residue cyclophane formation in the biosynthesis of a new family of RiPP natural products, the triceptides. This analysis revealed 3940 (3113 unique) putative precursor sequences predicted to be modified by 3-CyFEs. Several uncharacterized maturase systems were identified that encode unique precursor types. Functional studies were carried out in vivo in Escherichia coli to identify modified precursors containing His and Tyr residues. NMR analysis of the products revealed that Tyr and His can also be incorporated into cyclophane macrocycles by 3-CyFEs. Collectively, all aromatic amino acids can be incorporated by 3-CyFEs, and the cyclophane formation strictly occurs via a C(sp2)–C(sp3) cross-link between the (hetero)aromatic ring to Cβ. In addition to 3-CyFEs, we functionally validated an Fe(II)/α-ketoglutarate-dependent hydroxylase, resulting in β-hydroxylated residues within the cyclophane rings. This study reveals the potential breadth of triceptide precursors and a systematic approach for studying these enzymes to broaden the diversity of peptide macrocycles.
中文翻译:

三肽的生物合成景观揭示了在含有 Tyr 和 His 的基序上催化环芳形成的自由基 SAM 酶
肽衍生的环烷在大环肽的化学空间中占据了一个独特的位置,其中有几个具有药学重要性的例子。尽管合成和生物催化方法都可用于构建这些大环化合物,但能够掺入构成大环化合物的多种氨基酸的通用(生物)催化剂将可用于产生多种肽环烷。在本报告中,我们协同使用生物信息学工具来绘制自由基 SAM 酶 (3-CyFEs) 的生物合成图谱,这些酶在新的 RiPP 天然产物家族三肽的生物合成中催化三残基环烷的形成。该分析揭示了 3940 个(3113 个独特的)推定的前体序列,预计会被 3-CyFE 修饰。鉴定了几种编码独特前体类型的未表征的成熟酶系统。进行了功能研究在大肠杆菌中体内鉴定含有 His 和 Tyr 残基的修饰前体。产物的 NMR 分析表明 Tyr 和 His 也可以通过 3-CyFE 结合到环烷大环中。总的来说,所有芳香族氨基酸都可以通过 3-CyFE 结合,环烷的形成严格地通过(杂)芳香环与 Cβ 之间的 C(sp 2 )–C(sp 3 ) 交联发生。除了 3-CyFEs,我们还在功能上验证了一种 Fe(II)/α-酮戊二酸依赖性羟化酶,在环芳环内产生 β-羟基化残基。这项研究揭示了三肽前体的潜在广度以及研究这些酶以拓宽肽大环化合物多样性的系统方法。
更新日期:2022-06-21
中文翻译:

三肽的生物合成景观揭示了在含有 Tyr 和 His 的基序上催化环芳形成的自由基 SAM 酶
肽衍生的环烷在大环肽的化学空间中占据了一个独特的位置,其中有几个具有药学重要性的例子。尽管合成和生物催化方法都可用于构建这些大环化合物,但能够掺入构成大环化合物的多种氨基酸的通用(生物)催化剂将可用于产生多种肽环烷。在本报告中,我们协同使用生物信息学工具来绘制自由基 SAM 酶 (3-CyFEs) 的生物合成图谱,这些酶在新的 RiPP 天然产物家族三肽的生物合成中催化三残基环烷的形成。该分析揭示了 3940 个(3113 个独特的)推定的前体序列,预计会被 3-CyFE 修饰。鉴定了几种编码独特前体类型的未表征的成熟酶系统。进行了功能研究在大肠杆菌中体内鉴定含有 His 和 Tyr 残基的修饰前体。产物的 NMR 分析表明 Tyr 和 His 也可以通过 3-CyFE 结合到环烷大环中。总的来说,所有芳香族氨基酸都可以通过 3-CyFE 结合,环烷的形成严格地通过(杂)芳香环与 Cβ 之间的 C(sp 2 )–C(sp 3 ) 交联发生。除了 3-CyFEs,我们还在功能上验证了一种 Fe(II)/α-酮戊二酸依赖性羟化酶,在环芳环内产生 β-羟基化残基。这项研究揭示了三肽前体的潜在广度以及研究这些酶以拓宽肽大环化合物多样性的系统方法。


























 京公网安备 11010802027423号
京公网安备 11010802027423号