当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Protein–Nucleic Acid Interactions for RNA Polymerase II Elongation Factors by Molecular Dynamics Simulations
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-06-10 , DOI: 10.1021/acs.jcim.2c00121 Adan Gallardo 1 , Brandon M Bogart 1 , Bercem Dutagaci 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-06-10 , DOI: 10.1021/acs.jcim.2c00121 Adan Gallardo 1 , Brandon M Bogart 1 , Bercem Dutagaci 1
Affiliation
|
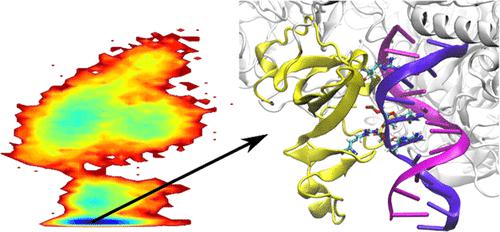
RNA polymerase II (Pol II) forms a complex with elongation factors to proceed to the elongation stage of the transcription process. In this work, we studied the elongation factor SPT5 and explored the protein–nucleic acid interactions for the isolated systems of KOW1 and KOW4 domains of SPT5 with DNA and RNA, respectively. We performed molecular dynamics (MD) simulations using three commonly used force fields that are CHARMM c36m, AMBER ff14sb, and ff19sb. Simulations showed strong protein–nucleic acid interactions and low electrostatic binding free energies for all force fields used. RNA was found to be highly dynamic with all force fields, while DNA had relatively more stable conformations with the AMBER force fields compared to that with CHARMM. Furthermore, we performed MD simulations of the complete elongation complex using CHARMM c36m and AMBER ff19sb force fields to compare the dynamics and interactions with the isolated systems. Similarly, strong KOW1 and DNA interactions were observed in the complete elongation complex simulations and DNA was further stabilized by a network of interactions involving SPT5–KOW1, SPT4, and rpb2 of Pol II. Overall, our study showed that the differences between CHARMM and AMBER force fields strongly affect the dynamics of the nucleic acids. CHARMM provides highly flexible DNA, while AMBER largely stabilizes the DNA structure. Although the presence of the entire interaction network stabilized the DNA and decreased the differences in the results from the two force fields, the discrepancies of the force fields for smaller systems may reflect their problems in generating accurate dynamics of nucleic acids.
中文翻译:

分子动力学模拟的 RNA 聚合酶 II 延伸因子的蛋白质-核酸相互作用
RNA 聚合酶 II (Pol II) 与延伸因子形成复合物,进入转录过程的延伸阶段。在这项工作中,我们研究了延伸因子 SPT5,并分别探索了 SPT5 的 KOW1 和 KOW4 结构域与 DNA 和 RNA 的分离系统的蛋白质-核酸相互作用。我们使用 CHARMM c36m、AMBER ff14sb 和 ff19sb 三个常用力场进行了分子动力学 (MD) 模拟。模拟显示,对于所有使用的力场,蛋白质-核酸相互作用强,静电结合自由能低。发现 RNA 在所有力场中都具有高度动态性,而与 CHARMM 相比,在 AMBER 力场中 DNA 具有相对更稳定的构象。此外,我们使用 CHARMM c36m 和 AMBER ff19sb 力场对完整的伸长复合物进行了 MD 模拟,以比较动力学和与孤立系统的相互作用。同样,在完整的延伸复合物模拟中观察到强烈的 KOW1 和 DNA 相互作用,并且 DNA 通过涉及 SPT5-KOW1、SPT4 和 Pol II 的 rpb2 的相互作用网络进一步稳定。总体而言,我们的研究表明,CHARMM 和 AMBER 力场之间的差异强烈影响核酸的动力学。CHARMM 提供高度灵活的 DNA,而 AMBER 在很大程度上稳定了 DNA 结构。尽管整个相互作用网络的存在稳定了 DNA 并减少了两个力场结果的差异,
更新日期:2022-06-10
中文翻译:

分子动力学模拟的 RNA 聚合酶 II 延伸因子的蛋白质-核酸相互作用
RNA 聚合酶 II (Pol II) 与延伸因子形成复合物,进入转录过程的延伸阶段。在这项工作中,我们研究了延伸因子 SPT5,并分别探索了 SPT5 的 KOW1 和 KOW4 结构域与 DNA 和 RNA 的分离系统的蛋白质-核酸相互作用。我们使用 CHARMM c36m、AMBER ff14sb 和 ff19sb 三个常用力场进行了分子动力学 (MD) 模拟。模拟显示,对于所有使用的力场,蛋白质-核酸相互作用强,静电结合自由能低。发现 RNA 在所有力场中都具有高度动态性,而与 CHARMM 相比,在 AMBER 力场中 DNA 具有相对更稳定的构象。此外,我们使用 CHARMM c36m 和 AMBER ff19sb 力场对完整的伸长复合物进行了 MD 模拟,以比较动力学和与孤立系统的相互作用。同样,在完整的延伸复合物模拟中观察到强烈的 KOW1 和 DNA 相互作用,并且 DNA 通过涉及 SPT5-KOW1、SPT4 和 Pol II 的 rpb2 的相互作用网络进一步稳定。总体而言,我们的研究表明,CHARMM 和 AMBER 力场之间的差异强烈影响核酸的动力学。CHARMM 提供高度灵活的 DNA,而 AMBER 在很大程度上稳定了 DNA 结构。尽管整个相互作用网络的存在稳定了 DNA 并减少了两个力场结果的差异,


























 京公网安备 11010802027423号
京公网安备 11010802027423号