Environment International ( IF 11.8 ) Pub Date : 2022-06-03 , DOI: 10.1016/j.envint.2022.107332 Jianfei Chen 1 , Yuyin Yang 2 , Yanchu Ke 1 , Xiuli Chen 1 , Xinshu Jiang 3 , Chao Chen 4 , Shuguang Xie 1
|
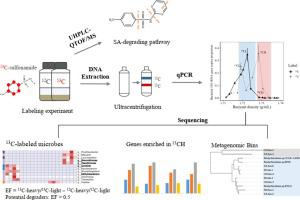
Sulfonamide (SA) antibiotics are ubiquitous pollutants in livestock breeding and aquaculture wastewaters, which increases the propagation of antibiotic resistance genes. Microbes with the ability to degrade SA play important roles in SA dissipation, but their diversity and the degradation mechanism in the field remain unclear. In the present study, we employed DNA-stable isotope probing (SIP) combined with metagenomics to explore the active microorganisms and mechanisms of SA biodegradation in antibiotic-contaminated wetland sediments. DNA-SIP revealed various SA-assimilating bacteria dominated by members of Proteobacteria, such as Bradyrhizobium, Gemmatimonas, and unclassified Burkholderiaceae. Both sulfadiazine and sulfamethoxazole were dissipated mainly through the initial ipso-hydroxylation, and were driven by similar microbes. sadA gene, which encodes an NADH-dependent monooxygenase, was enriched in the 13C heavy DNA, confirming its catalytic capacity for the initial ipso-hydroxylation of SA in sediments. In addition, some genes encoding dioxygenases were also proposed to participate in SA hydroxylation and aromatic ring cleavage based on metagenomics analysis, which might play an important role in SA metabolism in the sediment ecosystem when Proteobacteria was the dominant active bacteria. Our work elucidates the ecological roles of uncultured microorganisms in their natural habitats and gives a deeper understanding of in-situ SA biodegradation mechanisms.
中文翻译:

稳定同位素探测和宏基因组学揭示抗生素污染湿地沉积物中磺胺类代谢微生物及其机制
磺胺类抗生素是畜牧养殖和水产养殖废水中普遍存在的污染物,会增加抗生素抗性基因的传播。具有降解SA能力的微生物在SA消散中起着重要作用,但它们的多样性和现场降解机制仍不清楚。在本研究中,我们采用 DNA 稳定同位素探测 (SIP) 结合宏基因组学来探索抗生素污染湿地沉积物中 SA 生物降解的活性微生物和机制。DNA-SIP 揭示了以变形菌属成员为主的各种 SA 同化细菌,如慢生根瘤菌属、宝石单胞菌属和未分类的伯克霍尔德菌科。磺胺嘧啶和磺胺甲恶唑都主要通过最初的同羟基化消散,并由相似的微生物驱动。编码 NADH 依赖性单加氧酶的sadA基因在13 C 重 DNA 中富集,证实了其对初始ipso的催化能力- 沉积物中 SA 的羟基化。此外,基于宏基因组学分析,还提出了一些编码双加氧酶的基因参与了SA羟基化和芳环裂解,这可能在变形杆菌为主要活性细菌时在沉积物生态系统中的SA代谢中起重要作用。我们的工作阐明了未培养微生物在其自然栖息地中的生态作用,并加深了对原位SA 生物降解机制的理解。



























 京公网安备 11010802027423号
京公网安备 11010802027423号