Cell Chemical Biology ( IF 8.6 ) Pub Date : 2022-06-02 , DOI: 10.1016/j.chembiol.2022.05.005 Lu Xiao 1 , Linglan Fang 1 , Eric T Kool 1
|
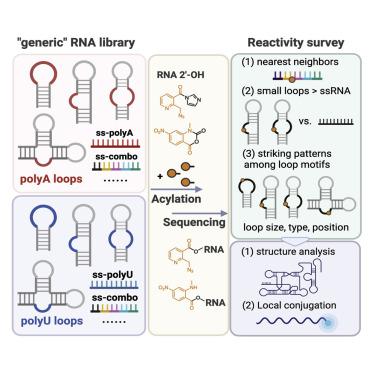
The reactivity of RNA 2ʹ-OH acylation is broadly useful both in probing structure and in preparing conjugates. To date, this reactivity has been analyzed in limited sets of biological RNA sequences, leaving open questions of how reactivity varies inherently without regard to sequence in structured contexts. We constructed and probed “generic” structured RNA libraries using homogeneous loop sequences, employing deep sequencing to carry out a systematic survey of reactivity. We find a wide range of RNA reactivities among single-stranded sequences, with nearest neighbors playing substantial roles. Remarkably, certain small loops are found to be far more reactive on average (up to 4,000-fold) than single-stranded RNAs, due to conformational constraints that enhance reactivity. Among loops, we observe large variations in reactivity based on size, type, and position. The results lend insights into RNA designs for achieving high-efficiency local conjugation and provide new opportunities to refine structure analysis.
中文翻译:

“通用”RNA 文库的酰化探测揭示了环限制对反应性的关键影响
RNA 2ʹ-OH 酰化的反应性在探测结构和制备缀合物方面具有广泛的用途。迄今为止,这种反应性已经在有限的生物 RNA 序列中进行了分析,留下了一个悬而未决的问题:反应性如何内在地变化,而不考虑结构化环境中的序列。我们使用均质环序列构建并探索了“通用”结构 RNA 文库,采用深度测序对反应性进行系统调查。我们发现单链序列之间存在广泛的 RNA 反应性,其中最近的邻居发挥着重要作用。值得注意的是,由于增强反应性的构象限制,某些小环的平均反应性比单链 RNA 高得多(高达 4,000 倍)。在环中,我们观察到基于大小、类型和位置的反应性存在很大差异。这些结果为实现高效局部缀合的 RNA 设计提供了见解,并为完善结构分析提供了新的机会。



























 京公网安备 11010802027423号
京公网安备 11010802027423号