当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
PointSite: A Point Cloud Segmentation Tool for Identification of Protein Ligand Binding Atoms
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-05-27 , DOI: 10.1021/acs.jcim.1c01512 Xu Yan 1 , Yingfeng Lu 1 , Zhen Li 1 , Qing Wei 1 , Xin Gao 2 , Sheng Wang 3, 4 , Song Wu 5 , Shuguang Cui 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-05-27 , DOI: 10.1021/acs.jcim.1c01512 Xu Yan 1 , Yingfeng Lu 1 , Zhen Li 1 , Qing Wei 1 , Xin Gao 2 , Sheng Wang 3, 4 , Song Wu 5 , Shuguang Cui 1
Affiliation
|
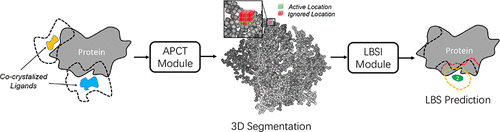
Accurate identification of ligand binding sites (LBS) on a protein structure is critical for understanding protein function and designing structure-based drugs. As the previous pocket-centric methods are usually based on the investigation of pseudo-surface-points outside the protein structure, they cannot fully take advantage of the local connectivity of atoms within the protein, as well as the global 3D geometrical information from all the protein atoms. In this paper, we propose a novel point clouds segmentation method, PointSite, for accurate identification of protein ligand binding atoms, which performs protein LBS identification at the atom-level in a protein-centric manner. Specifically, we first transfer the original 3D protein structure to point clouds and then conduct segmentation through Submanifold Sparse Convolution based U-Net. With the fine-grained atom-level binding atoms representation and enhanced feature learning, PointSite can outperform previous methods in atom Intersection over Union (atom-IoU) by a large margin. Furthermore, our segmented binding atoms, that is, atoms with high probability predicted by our model can work as a filter on predictions achieved by previous pocket-centric approaches, which significantly decreases the false-positive of LBS candidates. Besides, we further directly extend PointSite trained on bound proteins for LBS identification on unbound proteins, which demonstrates the superior generalization capacity of PointSite. Through cascaded filter and reranking aided by the segmented atoms, state-of-the-art performance can be achieved over various canonical benchmarks, CAMEO hard targets, and unbound proteins in terms of the commonly used DCA criteria.
中文翻译:

PointSite:用于识别蛋白质配体结合原子的点云分割工具
准确识别蛋白质结构上的配体结合位点 (LBS) 对于了解蛋白质功能和设计基于结构的药物至关重要。由于以前的以口袋为中心的方法通常基于对蛋白质结构外的伪表面点的研究,它们不能充分利用蛋白质内部原子的局部连通性以及来自所有结构的全局 3D 几何信息。蛋白质原子。在本文中,我们提出了一种新的点云分割方法PointSite,用于准确识别蛋白质配体结合原子,以蛋白质为中心的方式在原子水平上进行蛋白质LBS识别。具体来说,我们首先将原始的 3D 蛋白质结构转移到点云中,然后通过基于子流形稀疏卷积的 U-Net 进行分割。凭借细粒度的原子级绑定原子表示和增强的特征学习,PointSite 可以大大优于原子交叉联合(atom-IoU)中的先前方法。此外,我们的分段结合原子,即我们的模型预测的具有高概率的原子,可以作为先前以口袋为中心的方法实现的预测的过滤器,这显着降低了 LBS 候选者的假阳性。此外,我们进一步直接扩展了在结合蛋白上训练的 PointSite,用于对未结合蛋白进行 LBS 识别,这证明了 PointSite 卓越的泛化能力。通过分段原子辅助的级联过滤和重新排序,可以在各种规范基准、CAMEO 硬目标、
更新日期:2022-05-27
中文翻译:

PointSite:用于识别蛋白质配体结合原子的点云分割工具
准确识别蛋白质结构上的配体结合位点 (LBS) 对于了解蛋白质功能和设计基于结构的药物至关重要。由于以前的以口袋为中心的方法通常基于对蛋白质结构外的伪表面点的研究,它们不能充分利用蛋白质内部原子的局部连通性以及来自所有结构的全局 3D 几何信息。蛋白质原子。在本文中,我们提出了一种新的点云分割方法PointSite,用于准确识别蛋白质配体结合原子,以蛋白质为中心的方式在原子水平上进行蛋白质LBS识别。具体来说,我们首先将原始的 3D 蛋白质结构转移到点云中,然后通过基于子流形稀疏卷积的 U-Net 进行分割。凭借细粒度的原子级绑定原子表示和增强的特征学习,PointSite 可以大大优于原子交叉联合(atom-IoU)中的先前方法。此外,我们的分段结合原子,即我们的模型预测的具有高概率的原子,可以作为先前以口袋为中心的方法实现的预测的过滤器,这显着降低了 LBS 候选者的假阳性。此外,我们进一步直接扩展了在结合蛋白上训练的 PointSite,用于对未结合蛋白进行 LBS 识别,这证明了 PointSite 卓越的泛化能力。通过分段原子辅助的级联过滤和重新排序,可以在各种规范基准、CAMEO 硬目标、



























 京公网安备 11010802027423号
京公网安备 11010802027423号